1378
Towards Informative Uncertainty Measures for MRI Segmentation in Clinical Practice: Application to Multiple Sclerosis1Radiology department, Lausanne University Hospital (CHUV), Lausanne, Switzerland, 2Doctoral School of the Faculty of Biology and Medicine, University of Lausanne (UNIL), Lausanne, Switzerland, 3University of Applied Sciences of Western Switzerland, Sierre, Switzerland, 4University of Cambridge, Cambridge, United Kingdom, 5Icahn School of Medicine at Mount Sinai, New York, NY, United States, 6Shifts Project, Helsinki, Finland, 7University Hospital Basel, Basel, Switzerland, 8IBM Research Europe, Zurich, Switzerland, 9Center for Biomedical Imaging (CIBM), University of Lausanne, Lausanne, Switzerland
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Multiple Sclerosis, Machine learning/Artificial intelligence, Brain, Uncertainty estimation, Reliable AI
We approach the problem of quantifying the degree of reliability of supervised deep learning models used by clinicians for automatic multiple sclerosis lesion segmentation on MRI. In particular, we quantify the correspondence of various uncertainty measures to the errors that a deep learning model makes in overall segmentation or lesion detection. The evaluation is done both on in- and out-of- domain datasets (40 and 99 patients respectively), and provides insights about the measures that can point clinicians to potential errors of an automatic algorithm regardless of the distributional shift.Introduction
MRI plays an important role in diagnosing and monitoring multiple sclerosis (MS)1. White matter lesions (WML) identified on T2 and FLAIR brain scans is a hallmark of the disease1-3. Over the past years various deep learning (DL) algorithms have been developed to replace a time-consuming skill-demanding procedure of manual WML annotation4. On the other hand, WML segmentation with black-box DL models is not necessarily reliable, especially when tested on out-of-domain data, e.g. different scanners, centres, patients, etc6-9. Thus, automatic predictions should be verified and corrected by clinicians. In this work, we investigate different voxel- and lesion-scale uncertainty measures as a method of pointing clinicians to potential model errors in overall segmentation or lesion detection.Methods
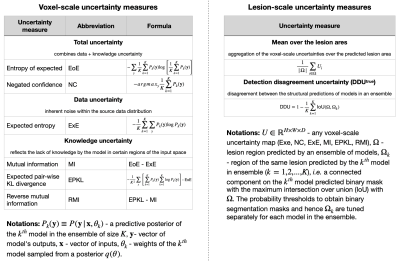
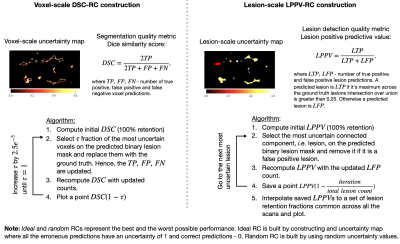
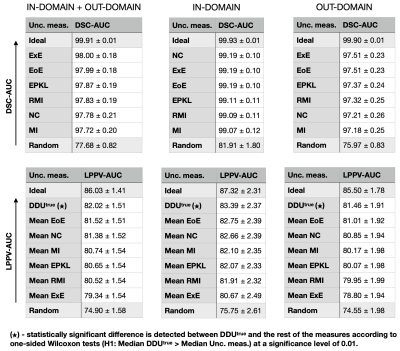
We evaluate six voxel-scale uncertainty measures6,9 and seven lesion-scale measures6-8(full list in Figure 1). Uncertainty is estimated using deep ensembles5, where the base model is a 3D U-net, which was previously used in uncertainty studies for the WML segmentation task6-9.The absolute values of uncertainty are not necessarily meaningful, hence we should only rely on the ranking of the uncertainties for different predictions. Error retention curves (RC) allow quantifying the correspondence between an uncertainty measure and model errors while only looking at the ranking of predictions in terms of uncertainty5,6. An RC for a single subject is built by iteratively replacing a fraction of the most uncertain predictions (voxels or lesions) with the ground truth, and recomputing model performance on this subject in terms of overall segmentation or lesion detection (see Figure 2). As a segmentation quality measure at the voxel scale the Dice similarity coefficient (DSC) is used; at the lesion scale the detection quality is evaluated using the lesion positive predictive value (LPPV) (see Figure 2). Average across subjects areas under respective RCs, i.e DSC-AUC or LPPV-AUC, quantify for the particular dataset the correspondence between voxel or lesion uncertainty measures and errors made in segmentation or lesion detection.
We employ a dataset provided by the Shifts project9. It contains FLAIR scans, which underwent denoising, skull stripping, bias field correction and interpolation to 1 mm3 space, and their manual WML annotations used as the ground truth. The Shifts dataset embraces four publicly available and one private datasets acquired at six different medical centres with six different scanner models (both 1.5T and 3T field strength). Training and validation sets contain data from four different medical centres with 33 and 7 scans respectively. The Shifts dataset allows to separate the RC analysis between in-domain (same centres as the training data) and out-of-domain (two new centres) sets containing 40 and 99 subjects respectively.
Results and Discussion
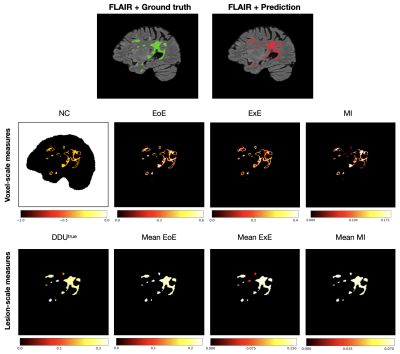
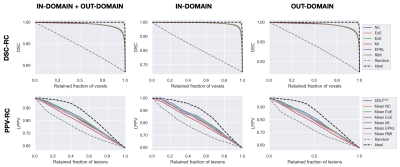
Examples of uncertainty maps on voxel and lesion scales are shown in Figure 3. The resulting voxel- and lesion-scale RCs computed separately for in- and out-of-domain data, as well as for the whole dataset are shown in Figure 4. The respective areas under the RCs are ranked and shown in Figure 5.The entropy based measures (ExE and EoE) have the highest DSC-AUC on the shifted dataset, indicating a superior ability in capturing model segmentation errors compared to other voxel-scale measures. However ExE loses informativeness for the lesion detection, showing the lowest LPPV-AUC. In principle, regions of high voxel uncertainty are often located on lesion borders and should be related to lesion delineation more than detection (Figure 3). The lesion-scale measure DDUtrue is not based on the voxel-scale uncertainty but computes the disagreement in structural predictions between models in an ensemble. DDUtrue shows the highest LPPV-AUC on both in- and out-of-domain data. Despite that, a visual examination of voxel uncertainty maps sometimes shows non-zero uncertainties inside false negative (FN) lesions, while lesion-scale uncertainties cannot be computed for FN lesions and, thus, cannot be used for FN localisation (see Figure 3).
On the other hand, the ranking of the voxel-scale uncertainty measures in terms of DSC-AUC is different for the in- and out-of-domain datasets. In particular, the DSC-AUC of the negated confidence measure is the highest in the initial domain, but is one of the lowest in the shifted domain. The ranking of the lesion uncertainty measures, however, does not change under the distributional shift.
Conclusions
In this study, we promote the use of uncertainty measures to quantify the degree of reliability of DL models for WML segmentation in MS. We compared different uncertainty measures both on voxel and lesion scales on the in- and out-of-domain data, showing that lesion-scale uncertainty measures in comparison to the voxel-scale ones yield a more consistent ranking of measures in terms of capturing model errors. Additionally, we observe that the lesion uncertainty DDUtrue has a superior ability to capture model errors related to lesion detection, what withholds for both in- and out-of-domain. We believe that lesion-scale detection uncertainty is needed to support the adoption of automatic DL-based methods for WML segmentation into the clinical practice. Our study guides towards which uncertainty measures are more informative for pinpointing potential errors in voxel- or lesion-scale predictions. It is yet important to verify in practice if the information brought by the uncertainty maps can simplify or speed up a semi-automatic segmentation by pointing clinicians to potential model errors.Acknowledgements
This work was supported by the Hasler Foundation Responsible AI programme (MSxplain) and the EU Horizon 2020 project AI4Media (grant 951911). We acknowledge access to the facilities and expertise of the CIBM Center for Biomedical Imaging, a Swiss research center of excellence founded and supported by Lausanne University Hospital (CHUV), University of Lausanne (UNIL), École polytechnique fédérale de Lausanne (EPFL), University of Geneva (UNIGE) and Geneva University Hospitals (HUG).References
1. Hemond CC, Bakshi R. Magnetic Resonance Imaging in Multiple Sclerosis. Cold Spring Harb Perspect Med. 2018;8(5):a028969. doi:10.1101/cshperspect.a028969
2. Thompson AJ, Banwell BL, Barkhof F, et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018;17(2):162-173. doi:10.1016/S1474-4422(17)30470-2
3. Bendfeldt K, Kuster P, Traud S, et al. Association of regional gray matter volume loss and progression of white matter lesions in multiple sclerosis - A longitudinal voxel-based morphometry study. Neuroimage. 2009;45(1):60-67. doi:10.1016/j.neuroimage.2008.10.006
4. Zeng C, Gu L, Liu Z, Zhao S. Review of Deep Learning Approaches for the Segmentation of Multiple Sclerosis Lesions on Brain MRI. Front Neuroinform. 2020;14:610967. Published 2020 Nov 20. doi:10.3389/fninf.2020.610967
5. Malinin A. Uncertainty estimation in deep learning with application to spoken language assessment, Ph.D. thesis, University of Cambridge, United Kingdom, 2019.
6. Molchanova N, Raina V, Malinin A, et al. Novel structural-scale uncertainty measures and error retention curves: application to multiple sclerosis. ArXiv.
7. Lambert B, Forbes F, Tucholka A, Doyle S, Dojat M. Multi-Scale Evaluation of Uncertainty Quantification Techniques for Deep Learning based MRI Segmentation. In ISMRM-ESMRMB & ISMRT 2022 - 31st Joint Annual Meeting International Society or Magnetic Resonance in Medicine London, United Kingdom, May 2022.
8. Nair T, Precu D, Arnold DL, Arbel T. Exploring Uncertainty Measures in Deep Networks for Multiple Sclerosis Lesion Detection and Segmentation. Medical image analysis. 2018,59:101557. doi:10.1007/978-3-030-00928-1_74
9. Malinin A, Athanasopoulos A, Barakovic M, et al. Shifts 2.0: Extending The Dataset of Real Distributional Shifts. ArXiv. doi:10.48550/arxiv.2206.15407
Figures