1369
A transfer learning approach to predict Axon Diameter and g-ratio distributions from MRI Data1Bioengineering Department, Stanford University, Stanford, CA, United States, 2Radiology Department, Stanford University, Stanford, CA, United States, 3Neurology Department, Stanford University, Stanford, CA, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Microstructure, Histology, Diffusion Imaging, g-ratio, axon diameter
To better establish the influence of histological features on the MRI signal, we present a multi-task neural network trained to predict parametrized microstructural distributions (axon diameters and g-ratios) from diffusion and magnetization transfer MRI data. To begin, we trained the model using histologically-derived synthetic MRI data before applying transfer learning by fine tuning on empirical data. Our initial results on both synthetic and empirical ex vivo mouse brain MRI data demonstrate the feasibility of this approach.Introduction
Different MRI contrasts such as Diffusion MRI (dMRI), Magnetization Transfer (MT), or relaxometry can be used to probe tissue microstructure non-invasively[1,2]. This is usually achieved by performing a voxel-wise fit of the MRI data to a biophysical model and then extracting parameters that represent specific microstructural features. Unfortunately, the biophysical models employed often rely on strong assumptions and can be prone to overfitting and misinterpretation[3]. Machine learning(ML) techniques have been proposed as a potential approach to learn the relationship between MRI and microstructural features[4-7]. However, a ground truth, obtained from ex vivo experiments that directly compare MRI and histology results in the same specimen[8,9], is typically required for training the model. Here, we present (i)an approach to train a neural network using synthetic MRI data derived from segmenting histological images and modeling the effects of the components on the MRI signal, (ii)a multi-task network to predict distributions of axon diameters and g-ratios, and (iii)transfer learning on empirical MRI data that demonstrate the potential of our approach.Methods
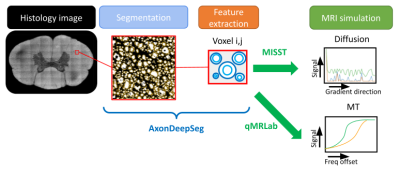
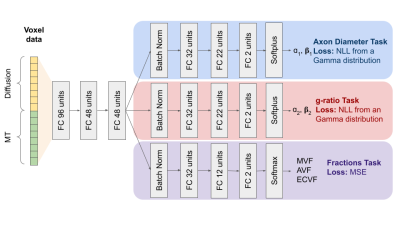
Electron Microscopy (EM) images were obtained from a publicly available dataset (https://osf.io/yp4qg/) which corresponds to single slices of canine spinal cord[10] and a rat spinal cord. MRI scans of the canine tissue sample were also included with this dataset and provided a reference to compare synthetic MRI data against. The method for generating histological-derived synthetic MRI is depicted in Figure 1. Histological images were divided into 150x150um(dog) and 50x50um(rat) tiles representing the regions that correspond to the MRI voxels. The axon and myelin within each tile were segmented using AxonDeepSeg[11] and AxonDeepSeg’s automatic labeling was used to obtain lists of g-ratios and axon diameters, which were fitted to a Gamma distribution(parameters: ɑ and ꞵ)[12,13]. Only tiles containing 400 or more axons were included. The segmentation was used to obtain ground truth myelin volume fraction(MVF), axon volume fraction(AVF) and extracellular volume fraction(ECVF). The dMRI was simulated using cylinders embedded in an isotropic compartment via the MISST toolbox[14]. The signals were derived from rectangular diffusion gradients with b-values of 3000 and 6000s/mm2, 35 encoding directions, five b=0s/mm2 and Δ/δ=18/5 ms. The MT images were simulated using qMRLab[15] based on the groundtruth MVF, accompanying R1 maps and physical constants were taken from the literature[16] using flip angles of 1000° and 4000°, and 7 offsets (1-30Khz). Rician noise was added to all simulations to obtain an SNR of 30dB. The acquisition protocol was chosen to match nine previously scanned mouse brains[17] on a 7T Bruker small bore animal MRI including 4 wildtype and 5 absence seizure mutant mice[18].For the neural network architecture, we chose a multi-task approach where a fully connected network performs three prediction tasks based on the raw MRI signal values for a given voxel: the parameters of two gamma distributions for the axons and g-ratios, and another branch that fits the MVF, AVF and ECVF (Figure 2). The axon and g-ratio branches are trained using a negative log-likelihood loss on the axon and g-ratio samples while for the fraction prediction we used a mean squared error loss with respect to the ground truth fractions. The network was programmed in Tensorflow[19,20]. Training was performed in 80% of the simulated data (~1200 voxels) during 3500 epochs with SGD and learning rate(LR) of 0.002. 10% of the data was used for validation and 10% for testing.
After the network was trained, we used segmented data from a region of the genu of 9 mouse brains MRI with manually annotated axons and g-ratios[21] and fractions extracted by AxonDeepSeg. We used one wildtype and one mutant mouse for training and the same quantity for validation to fine tune the network previously trained on simulated data (1000 epochs, Adam with LR=0.001, the first common layer and the batchnorm layer of the g-ratio branch were frozen). We report test results on the remaining 5 mouse brains.
Results and Discussion
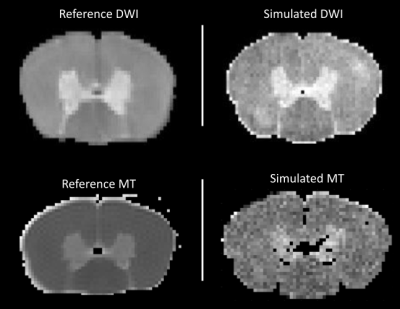
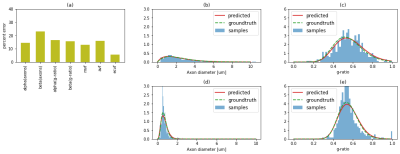
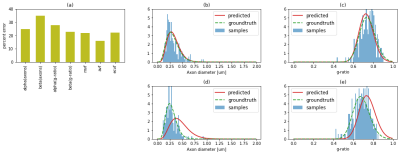
Figure 3 shows an example DWI and MT synthetic images compared to empirically acquired data in the same samples. Despite the fact that the synthetic data is based on subsampled histological features and some imperfections in the automatic segmentation, we observe a good visual agreement between the synthetic and empirical MRI[10]. Figure 4a shows simulations from the test-set with mean absolute errors ranging from 3 to 18%. Figure 4b-c, shows histograms of the groundtruth and predicted axon diameter and g-ratio distributions from the test-set. We observe a good agreement (<25% error in all predicted parameters) between the fitted groundtruth distribution and the ML prediction. Analogous plots for the experimental mouse data are shown in Figure 5. Here, we observe one mouse brain sample with good agreement between the predicted and ground truth histology (%error: axon diameters: ɑ=14.2%,ꞵ=9.1%; g-ratios: ɑ=12.7%,ꞵ=14.4%) and a second sample with higher percent error (%error: axon diameters: ɑ=16.9%,ꞵ=47.6%; g-ratios: ɑ=23.5%,ꞵ=13.6%) .Conclusion
We have presented histologically-derived synthetic MRI data that enabled training a multi-task ML approach to predict parametrized axon diameter and g-ratio distributions and microstructural fractions from raw MRI data. Our experiments show the potential of training using synthetic images and fine-tuning with empirical MRI data. Future work will focus on a wider range of synthetic data and further experimental testing.Acknowledgements
*These authors contributed equally to this work
This work was partially supported by the Stanford Wu Tsai Neurosciences Institute through an Stanford Interdisciplinary Graduate Fellowship and Stanford Enhancing Diversity in Graduate Education (EDGE) fellowship.
The authors would like to acknowledge the helpful discussions with Dr. Charles Huang regarding the architecture of the neural network.
References
1. Novikov, Dmitry S. "The present and the future of microstructure MRI: From a paradigm shift to normal science." Journal of Neuroscience Methods 351 (2021): 108947.
2. Weiskopf, Nikolaus, et al. "Quantitative magnetic resonance imaging of brain anatomy and in vivo histology." Nature Reviews Physics 3.8 (2021): 570-588.
3. A. Rokem, et al. "Evaluating the accuracy of diffusion mri models in white matter." PLOS ONE 10.4 (2015): 1 – 26 .
4. Liang, Zifei, et al. "Virtual mouse brain histology from multi-contrast MRI via deep learning." Elife 11 (2022): e72331.
5. Hédouin, Renaud, et al. "Decoding the microstructural properties of white matter using realistic models." NeuroImage 237 (2021): 118138.
6. E. Weber, C. Leuze, D. Barbosa, G. Chau, K. Grill-Spector and J. McNab, “Prediction ofhuman brain microstructure from raw diffusion MRI data using deep neural networks,” 2022 ISMRM Workshop on Ultra-High Field MR.
7. E. Weber, C. Leuze, D. Barbosa, G. Chau, K. Grill-Spector and J. McNab, "Learning the relationship between human brain tissue microstructure and diffusion MRI data" (2021) in: Proceedings of the 30th Annual Meeting of ISMRMA.
8. Seehaus et al. "Histological validation of high-resolution dti in human post mortem tissue." Frontiers in Neuroanatomy 9 (2015): 98.
9. K. G. Schilling et al. "Histological validation of diffusion mri fiber orientation distributions and dispersion." NeuroImage 165 (2018): 200 – 221.
10. Vuong, M.-T., Duval, T., Cohen-Adad, J., Stikov, N., 2017. On the Precision of Myelin Imaging: Characterizing Ex Vivo Dog Spinal Cord with MRI and Histology, in: Proceedings of the 25th Annual Meeting of ISMRM. p. 3760.
11. Zaimi, Aldo, et al. "AxonDeepSeg: automatic axon and myelin segmentation from microscopy data using convolutional neural networks." Scientific reports 8.1 (2018): 1-11.
12. Assaf, Yaniv, et al. "AxCaliber: a method for measuring axon diameter distribution from diffusion MRI." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 59.6 (2008): 1347-1354.
13. West, Kathryn L., et al. "A revised model for estimating g-ratio from MRI." Neuroimage 125 (2016): 1155-1158.
14. Ianuş, Andrada, Daniel C. Alexander, and Ivana Drobnjak. "Microstructure imaging sequence simulation toolbox." International workshop on simulation and synthesis in medical imaging. Springer, Cham, 2016.
15. Karakuzu, Agah, et al. "qMRLab: Quantitative MRI analysis, under one umbrella." Journal of Open Source Software 5.53 (2020): 2343.
16. West, Kathryn L., et al. "Myelin volume fraction imaging with MRI." Neuroimage 182 (2018): 511-521.
17. Chau Loo Kung, Gustavo, et al. "Myelin Alterations during the Development of an Absence Seizure Mouse Model." in: Proceedings of the 31st Annual Meeting of ISMRM.
18. Makinson, Christopher D., et al. "Regulation of thalamic and cortical network synchrony by Scn8a." Neuron 93.5 (2017): 1165-1179.
19. Martín Abadi., et al. TensorFlow: Large-scale machine learning on heterogeneous systems,2015. Software available from tensorflow.org.
20. Dillon, Joshua V., et al. "Tensorflow distributions." arXiv preprint arXiv:1711.10604 (2017).
21. Chau Loo Kung, Gustavo et al. “Validation of MRI Measurements of Myelination Changes in an Absence Epilepsy Mouse Model,” in: Proceedings of the 30th Annual Meeting of ISMRM.
Figures