1355
Towards a multi-parametric MRI-based myelin marker for detecting enhanced myelination in the Gli1-/- mouse brain1Bernard and Irene Schwartz Center for Biomedical Imaging, Department of Radiology, New York University School of Medicine, New York, NY, United States, 2Department of Neuroscience and Physiology, Neuroscience Institute, New York University School of Medicine, New York, NY, United States, 3UConn Health, Farmington, CT, United States
Synopsis
Keywords: Multi-Contrast, Microstructure
Although multiple MRI-based myelin marker have been introduced, their sensitivity and specificity remained limited. Multi-parametric MRI can potentially enhance myelin mapping, but validation remains challenging. In this study, we compared myelin histology from the Gli1-/- mouse brain, which has enhanced myelination, with MRI markers based on relaxation, magnetization transfer, and diffusion properties from the same animals. We found varying degrees of correlation between MRI markers and myelin basic protein signals in multiple brain regions. Partial least square regression analysis demonstrated that multi-parametric MRI can indeed improve myelin mapping and provided information for further optimization.Introduction
Myelin plays an important role in maintaining normal brain functions and is a central therapeutic target in primary de/dysmyelinating disorders. MRI provides an array of versatile tissue contrasts, including contrasts based on tissue relaxation (T1, T2), magnetization transfer (MT), and diffusion properties, for the sensitive detection of myelin injuries. The sensitivity and specificity of these MRI parameters for myelin, however, are not well understood due to the lack of a direct correspondence between MRI signals and myelin. In fact, a recent meta-analysis1 demonstrates that only a few MRI-based markers strongly correlate with histological measurements, and “there is no myelin MRI measure true to histology”. A promising approach to gain myelin specificity is to combine the strengths of multiple MRI contrasts, each targeting a distinct aspect of myelin, and utilize statistical or multi-parameter modeling methods2,3 to characterize myelin. For example, the myelin information shared by MT and T2* MRI signals can be extracted to improve the specificity to the cortical myelin4. However, collinearity among MRI parameters needs to be considered, and, more importantly, ground truth myelin histology is critical for validation.In this study, we acquired a multi-parametric MRI data from the Gli1-/- mouse brain, which shows enhanced myelination in the corpus callosum and cortical regions from recruited neural stem cells5. We used partial least square regression (PSLR), which can remove collinearity among MRI parameters, to find relationships between myelin basic protein (MBP) stained histology and MRI parameters.
Methods
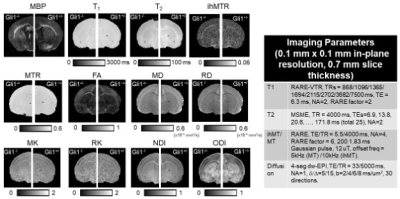
MRI: Ex vivo MRI data from Gli1+/- and Gli1-/- mouse brain (postnatal day 10 (P10), n = 3/3) were acquired using a 7 Tesla MRI system at room temperature. Co-registered T1, T2, magnetization transfer ratio (MTR), inhomogeneous MTR (ihMTR), and diffusion MRI metric maps (see Fig.1 for details) were acquired using the parameters shown in Fig. 1. MBP-stained histology was obtained from the same animals (20 um thickness, 20X). Analysis: Regions of interest (ROIs) were manually defined in the fractional anisotropy (FA) images and matching MBP-stained images for the genu and splenium of the corpus callosum (gcc and scc), cerebral peduncle (cp), external capsule (ec), motor and sensory cortex (mCX and sCX), and thalamus. Average MRI parameters and MBP intensities for each ROI were obtained.Results
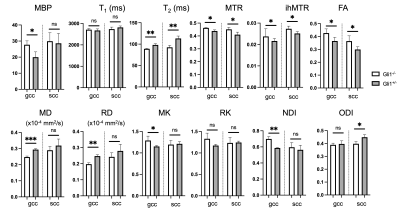
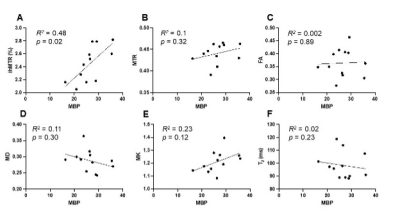
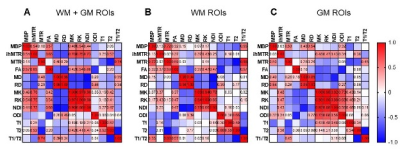
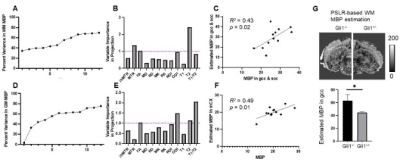
Multi-parametric MRI and histology of Gli1-/- mice: Fig. 1 shows representative MBP-stained histology and maps of eleven MRI parameters. The white matter (WM) and gray matter (GM) contrasts in MRI were relatively low due to low myelin content in the P10 mouse brain (myelination starts at around P7 in the mouse brain). As expected, the MBP values in the gcc of Gli1-/- mice were significantly higher than Gli1+/- mice (Fig. 2). Significant differences in the gcc between the Gli1-/- and Gli1+/- mice were found in ihMTR and several dMRI parameters (Fig. 2). However, only ihMTR in the gcc and scc correlated with MBP intensities (Fig. 3). Correlation matrices for measurements from multiple ROIs (Fig. 4) showed collinearity among MRI parameters.PSLR-based myelin predictor: PSLR uses latent components to remove collinearity among input parameters. PSLR results of WM ROI data showed that 9 components were needed to explain 70% variance in WM MBP signals (Fig. 5A), with major contributions from MTR, FA, and T2 (variable importance in projection > 1, Fig. 5B). In comparison, more than 10 components were needed to explain 70-80% variance in GM MBP signals (Fig. 5D), with major contributions from FA, ODI, T2, and T1/T2 (Fig. 5E). The PSLR-predicted MBP values from MRI parameters showed strong correlations with actual MBP signals (Fig. 5C and 5F), with significant differences between the Gli1-/- and Gli1+/- mice (Fig. 5G).
Discussion
Our results from the Gli1 mouse model are in line with previous reports1. For example, MTR showed a strong correlation with MBP signals in WM ROIs (Fig. 4B). It is interesting that ihMTR showed a strong correlation with MBP signals in the corpus callosum (Fig. 3) but relatively weak correlations with MBP signals from other WM ROIs (Fig. 4B), suggesting the relationship between them is complex. The PSLR results (Fig. 5) suggest that multiple MRI parameters are needed to account for the variances observed in tissue MBP signals, but choosing the optimal set of MRI parameters will require additional studies.This study has several limitations. Both the choice of ex vivo MRI, which is known to differ from in vivo MRI, and the early stage of myelination at P10, make it difficult to generalize the findings here to in vivo studies and mouse brains at later stages. In addition, there are other key aspects of myelin (e.g. myelin lipid) not captured by the MBP signals here, and several MRI myelin markers (e.g. myelin water fraction) were not included in this study. Future studies are necessary to further our understanding of optimal MRI-based myelin marker.
Conclusion
Multi-parameter MRI can enhance our ability to detect myelin and myelin repair.Acknowledgements
No acknowledgement found.References
1 Mancini, M. et al. An interactive meta-analysis of MRI biomarkers of myelin. Elife 9, doi:10.7554/eLife.61523 (2020).
2 Knight, M. J., Wearn, A., Coulthard, E. & Kauppinen, R. A. T2 Relaxometry and Diffusion Tensor Indices of the Hippocampus and Entorhinal Cortex Improve Sensitivity and Specificity of MRI to Detect Amnestic Mild Cognitive Impairment and Alzheimer's Disease Dementia. J Magn Reson Imaging 49, 445-455, doi:10.1002/jmri.26195 (2019).
3 Cercignani, M. & Bouyagoub, S. Brain microstructure by multi-modal MRI: Is the whole greater than the sum of its parts? Neuroimage 182, 117-127, doi:10.1016/j.neuroimage.2017.10.052 (2018).
4 Mangeat, G., Govindarajan, S. T., Mainero, C. & Cohen-Adad, J. Multivariate combination of magnetization transfer, T2* and B0 orientation to study the myelo-architecture of the in vivo human cortex. Neuroimage 119, 89-102, doi:10.1016/j.neuroimage.2015.06.033 (2015).
5 Samanta, J. et al. Inhibition of Gli1 mobilizes endogenous neural stem cells for remyelination. Nature 526, 448-452, doi:10.1038/nature14957 (2015).
Figures