1333
Deep learning for automatic detection and contouring of parotid gland tumors on MRI1Department of Imaging and Interventional radiology, The Chinese University of Hong Kong, Prince of Wales Hospital, HongKong, China, 2Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, China
Synopsis
Keywords: Cancer, Head & Neck/ENT
Parotid gland tumors (PGTs) are often asymptomatic and an incidental finding on MRI that can be overlooked. We constructed an accurate artificial intelligence (AI) tool trained on fat-suppressed T2-weighted MRI to automatically identify patients with PGTs with an accuracy of 94.3% (99/105), a sensitivity of 94.0% (47/50) and a specificity of 94.5% (52/55). For identified PGT patients, automatic segmentations of the tumor and gland were performed and achieved dices of 77.2% and 86.3%, respectively. The proposed AI tool may assist radiologists by acting as a second pair of eyes to ensure incidental PGTs on MRI are not missed.Introduction
Parotid gland tumors (PGTs) are often asymptomatic and detected incidentally by imaging, such as MRI to stage head and neck cancer (1). However, the head and neck is a complex region, a full assessment is time-consuming, and it is easy to overlook incidental findings if the search is not meticulous. An imaging program, to automatically detect PGTs would be helpful by acting as a second pair of eyes to alert the radiologist to a tumor in the parotid gland (PG). Furthermore, once PGTs are identified, the same program could be then used to contour and map the extent of the tumor to aid in treatment planning. Deep convolutional neural networks (CNNs) (2,3) offer the possibility of consistent, objective, and highly efficient methods to automatically detect and segment these tumors. Nonetheless, automatically identifying and contouring PGTs is still challenging due to their variable shape, low contrast, and similar intensity with surrounding structures. It remains unknown if the CNNs can successfully automatically detect and contour PGTs on MRI. Moreover, most segmentation studies only focus on datasets containing individuals with lesions but not individuals without lesions, limiting widespread clinical application. Recently, a self-configurable CNN architecture nnUNet (4) has emerged as a state-of-the-art medical image segmentation architecture. Therefore, we constructed a nnUNet-based artificial intelligence (AI) tool embedded with the pre-processing and PG-specific post-processing to first, discriminate patients with PGTs from those without PGTs, and second for the identified PGT patients, contour the tumor and the normal PG to assist in treatment planning.Methods
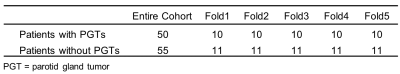
Collected DatasetThe dataset comprised 50 patients with PGTs and 55 patients without PGTs who underwent MRI with fat-suppressed T2-weighted imaging (FS-T2WI). The normal PGs and PGTs were manually drawn by an experienced head and neck researcher. Tumors with a maximum short diameter larger than 5mm were contoured, and this threshold is used to exclude small intraparotid nodes which are frequently found in normal PGs (5). All tumors were confirmed on cytology or histology.
Pre-processing and Augmentation Strategies
All images were first resampled to the same target spacing. Before training, the training data were cropped to their non-zero region and z-score normalization was adopted for each image. During training, augmentations such as rotation, scaling, random elastic deformation, mirroring, gaussian noise, gaussian blur, brightness and contrast adjustment, simulation of low resolution, and gamma intensity transform are adopted to each training sample.
Implementation Details
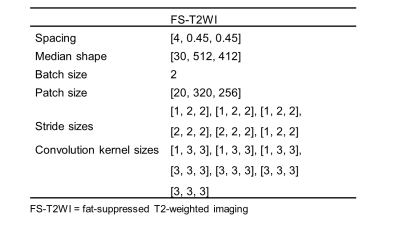
To avoid selection bias, 5-fold cross-validation was used to verify the model’s performance. All experiments were performed based on Python 3.6.13, PyTorch (Torch 1.8.1) and Ubuntu 18.04 on NVIDIA Tesla V100 32GB GPUs. The nnUNet architecture is adapted from 3D U-Net. It comprises a symmetrical encoder-decoder architecture with skip connections. The network configuration (Table 1) was automatically determined by nnUNet. The default optimizer is the Stochastic gradient descent algorithm, setting the momentum to 0.99. We trained 500 epochs ($$$\text{epoch}_{max}$$$ is 0.01) for each model. The learning rate ($$$\text{initial}_{lr}$$$) decreases by employing a poly learning rate policy:
$$lr = \text{initial}_{lr} \times \left(1-\frac{\text{epoch}_{id}}{\text{epoch}_{max}}\right)^{0.9}$$
Loss Function
To address the challenge caused by imbalanced data distribution, we employed a hybrid loss consisting of contributions from both dice loss and focal loss (6), which could learn the class distribution alleviating the imbalanced voxel problem. The total loss can be formulated as follow:
$$\mathcal{L} = \mathcal{L}_{Dice}\ + \lambda_{Focal}$$
Post-processing
Lesions with a maximum diameter of less than 5mm were removed to reduce false positive results from normal small parotid nodes in normal PGs.
Evaluation Metrics of the AI Detecting and Contouring Tool
Evaluation metrics for identifying patients with PGTs from those without PGTs include accuracy, sensitivity, and specificity. Evaluation metrics for comparing the AI-generated contour with manual contour include dice coefficient, Jaccard index, precision, and recall.
Results
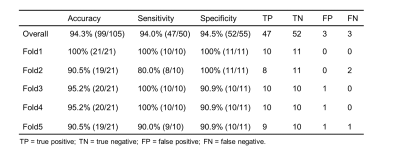
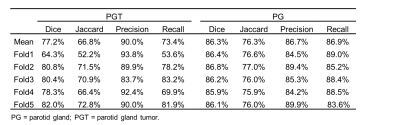
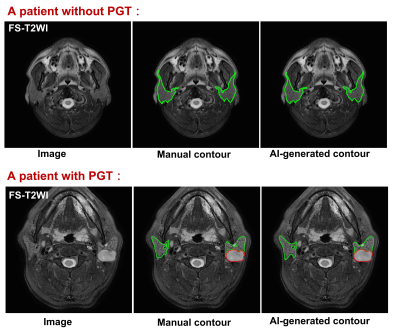
The dataset demographics of the entire cohorts are detailed in Table 2. For identifying patients with PGTs from patients without PGTs based on FS-T2WI (Table 3), our proposed AI tool achieved an overall accuracy of 94.3% (99/105), a sensitivity of 94.0% (47/50) and a specificity of 94.5% (52/55), respectively. For identified PGT patients, a comparison of the AI-generated contours with the manual contours produced mean dices of 77.2% and 86.3%, Jaccard indexes of 66.8% and 76.3%, precisions of 90.0% and 86.7%, recalls of 73.4% and 86.9% for tumors and normal PGs, respectively (Table 4). Examples of the patients with or without PGTs identified and contoured by our proposed AI tool are shown in Figure 1.Discussion
In this study, we constructed an AI detecting-contouring tool using a dataset of FS-T2WI MRI with or without PGTs. The proposed AI tool achieved high performances in identifying PGT patients and contouring the PGTs and normal PGs. Our study has several limitations, we only trained the AI tool on a small number of datasets and did not validate our AI tool with external data in this preliminary study. A larger data set need to be collected from a multicentre to circumvent this deficiency in the future.Conclusion
The proposed robust, accurate, efficient AI tool may assist radiologists in speeding up the detection and contouring of PGTs, and by acting as a second pair of eyes to ensure incidental PGTs on MRI are not missed.Acknowledgements
No acknowledgement found.References
1. Nam IC, Baek HJ, Ryu KH, Moon JI, Cho E, An HJ, Yoon S, Baik J. Prevalence and Clinical Implications of Incidentally Detected Parotid Lesions as Blind Spot on Brain MRI: A Single-Center Experience. Medicina-Lithuania 2021;57(8): 836.
2. Lin L, Dou Q, Jin YM, Zhou GQ, Tang YQ, Chen WL, Su BA, Liu F, Tao CJ, Jiang N, Li JY, Tang LL, Xie CM, Huang SM, Ma J, Heng PA, Wee JTS, Chua MLK, Chen H, Sun Y. Deep Learning for Automated Contouring of Primary Tumor Volumes by MRI for Nasopharyngeal Carcinoma. Radiology 2019;291(3):677-686.
3. Wong LM, Ai QYH, Mo FKF, Poon DMC, Ann DK. Convolutional neural network in nasopharyngeal carcinoma: how good is automatic delineation for primary tumor on a non‑contrast‑enhanced fat‑suppressed T2‑weighted MRI? Japanese Journal of Radiology, 39(6), 571-579.
4. Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18(2):203-211.
5. Zhang MH, Ginat DT. Normative measurements of parotid lymph nodes on CT imaging. Surg Radiol Anat 2020;42(9):1109-1112.
6. Lin TY, Goyal P, Girshick R, He KM, Dollar P. Focal Loss for Dense Object Detection. Ieee T Pattern Anal 2020;42(2):318-327.
Figures