1317
Similarity-driven motion-resolved reconstruction for ferumoxytol-enhanced whole-heart MRI of congenital heart disease patients1Diagnostic and Interventional Radiology, University Hospital of Lausanne (CHUV), Lausanne, Switzerland, 2Advanced Clinical Imaging Technology, Siemens Healthineers International AG, Lausanne, Switzerland, 3IHU LIRYC, Electrophysiology and Heart Modeling Institute, Université de Bordeaux – INSERM U1045, Pessac-Bordeaux, France, 4Department of Cardiovascular Imaging, Hôpital Cardiologique du Haut-Lévêque, CHU de Bordeaux, Pessac, France, 5INSA-Lyon, CNRS, Inserm, CREATIS, Université de Lyon, Villeurbanne, France, 6Département de Radiologie, Louis Pradel Hospital, Hospices Civils de Lyon, Bron, France, 7Woman-Mother-Child Department, Lausanne University Hospital and University of Lausanne, Division of Pediatric Cardiology, Lausanne, Switzerland, 8Heart and Vessel Department, Lausanne University Hospital and University of Lausanne, Service of Cardiology, Lausanne, Switzerland, 9Cardiac MR Center, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 10Center for Biomedical Imaging (CIBM), Lausanne, Switzerland
Synopsis
Keywords: Heart, Motion Correction
Ferumoxytol-enhanced free-running whole-heart MRI allows for a comprehensive evaluation of the cardiovascular anatomy in 3D. A similarity-driven multi-dimensional binning algorithm (SIMBA) has been proposed as a fast and efficient reconstruction of such data, by clustering and selecting motion-consistent information. In this work, we extend the SIMBA reconstruction to make use of the inherent redundancy of motion-consistent information using a compressed-sensing reconstruction, in which sparsity is maximized by the integration of inter-cluster non-rigid 3D motion-fields. With this new framework we demonstrate improved image quality, increased coronary sharpness and vessel conspicuity.
Introduction
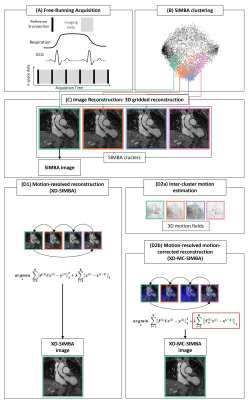
Ferumoxytol-enhanced whole-heart MRI enables accurate evaluation of the whole 3D cardiac anatomy, including origin and course of the coronary arteries in patients with congenital heart disease (CHD)1. In combination with free-running acquisitions2 it allows for robust self-gated physiological signal extraction and dynamic XD-GRASP 3D reconstructions3. A computationally efficient, static reconstruction of the same free-running data can be obtained using a similarity-driven multi-dimensional binning algorithm (SIMBA)4, without any assumptions on physiology. With SIMBA, though multiple motion-consistent clusters are obtained, only one is selected for image reconstruction. The goal of this work is to explore how SIMBA can be extended to use the redundant information that is shared among the clusters. In pursuit of this goal, we address two hypotheses: First, that the motion-consistent clusters can become a new dynamic dimension as part of a compressed sensing (CS) reconstruction. Second, that since predicting specific physiological states using SIMBA is not possible, the sparsity of the clustering dimension may be patient-dependent (e.g., with systolic clusters close to diastolic clusters) such that the direct application of CS can lead to suboptimal image quality. Therefore we test that non-rigid inter-cluster motion-field registration integrated into CS leads to improved image quality.Methods
Datasets from 27 CHD patients (1-60yo, 23$$$\pm$$$16kg, 20 males) were acquired on a 1.5T clinical MRI scanner (MAGNETOM Sola, Siemens Healthcare, Erlangen, Germany), after injection of ferumoxytol (2-4mg/kg) using a free-running GRE research application sequence with a 3D radial phyllotaxis trajectory5. SIMBA was applied as in 4 to obtain a set of motion-consistent clusters. While the original reconstruction (SIMBA) only takes the largest cluster of data for image reconstruction, by selecting the four most populated motion-consistent clusters, we exploit this redundant information using a CS reconstruction6. Therefore, we perform a motion-resolved reconstruction that takes the reconstructed SIMBA clusters as motion states (XD-SIMBA). We correct for potentially large deformations by estimating 3D non-rigid motion-fields7 between pairs of adjacent clusters, which are incorporated into the reconstruction framework (XD-MC-SIMBA) (Figure 1). The new reconstruction problem can be formulated as:$$$\sum_{i=1}^K\parallel F^{(i)}Cx^{(i)}-y^{(i)}\parallel^2_{2}+\lambda\sum_{i=1}^K\parallel\mathcal{T}_u^{(i)}x^{(i)}-x^{(i-1)}\parallel_{1}$$$
where $$$F$$$ is the non-uniform fast Fourier transform NUFFT8, $$$C$$$ the coil sensitivities, $$${x^{(i)}}$$$ the image reconstructed from the cluster $$$i$$$ where cyclical motion was enforced by setting $$$x^{(0)}=x^{(K)}$$$, $$$y^{(i)}$$$ the k-space data in the cluster $$$i$$$, $$$K=4$$$ the number of clusters taken, and $$$\mathcal{T}_u^{(i)}$$$ the image deformation operator applying the non-rigid motion-fields $$$u$$$ from the image $$$x^{(i)}$$$ to the reference image $$$x^{(i-1)}$$$. The regularization parameter $$$\lambda$$$ was experimentally set to 0.3. This problem was solved via operator-splitting using the alternating direction method of multipliers (ADMM).
To objectively compare the quality of the reconstructions, we evaluated: the contrast between blood and myocardium, the sharpness of the lung-liver and blood-myocardium interfaces9, the sharpness of the first 2cm of the right coronary artery (RCA) and the combined left main (LM) and left anterior descending coronary artery (LAD) using the Soap-Bubble tool10. Image quality scores (IQS), ranging from 0 (non-diagnostic) to 4 (excellent-diagnostic value), were assigned to each 3D volume by using a deep-learning-based approach11. The percentage of the acquired data used for each reconstruction was reported. Statistical analyses of all metrics were performed using one-way analysis of variance (ANOVA) with Bonferroni correction. Statistical significance was defined by two-sided paired sample t-tests with p<0.05.
Results
On average, SIMBA uses 12$$$\pm$$$1.9% of the acquired data, while XD-SIMBA and XD-MC-SIMBA 41$$$\pm$$$4.4%.Two representative ferumoxytol-enhanced 3D whole-heart datasets are shown in Figure 2. For both patients, all three reconstruction techniques allow a good visualization of the congenital defect, while the XD-MC-SIMBA image has the highest conspicuity.
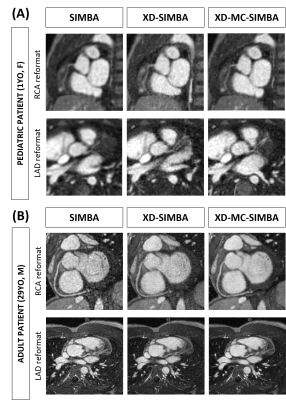
In Figure 3, LAD and RCA reformats are displayed for two CHD patients. These two examples demonstrate how XD-SIMBA can lead to an inferior visualization of the coronary arteries in terms of vessel conspicuity, while with XD-MC-SIMBA vessel visibility and sharpness are improved, particularly for the more distal segments.
Both lung-liver and blood-myocardium sharpness improvements are statistically significant for XD-MC-SIMBA (lung-liver: p=0.01; blood-myocardium: p=0.02; Figure 4A). Conversely, the blood-myocardium contrast ratio does not show significant differences (p=0.43, Figure 4A). Analyses of the coronary arteries (Figure 4B) suggest a statistically non-significant trend for an increased visible vessel length in XD-MC-SIMBA for both LM+LAD and RCA compared to SIMBA and XD-SIMBA (LM+LAD: p=0.16; RCA: p=0.19). The LAD sharpness is very similar between SIMBA and XD-MC-SIMBA while reduced for XD-SIMBA. For the RCA, vessel sharpness is highest for XD-MC-SIMBA (p=0.46).
The IQS comparison (Figure 4C) demonstrates how XD-MC-SIMBA leads to an improved image quality on average.
Consistent with these numerical findings, Figure 5 shows an example of much improved anatomical visualization of the aortic valve and coronary arteries with XD-MC-SIMBA when compared to both SIMBA and XD-SIMBA.
Discussion and Conclusions
In this work, we developed an improved SIMBA reconstruction (XD-MC-SIMBA) that better exploits the inherent abundancy of information from a free-running acquisition by using the SIMBA clustering as a new dimension of sparsity for CS reconstruction. When combined with a non-rigid inter-cluster motion-field registration, we conclude that XD-MC-SIMBA leads to improved image quality in a cohort of CHD patients. Investigations on the ideal and subject-specific number of clusters chosen and their ordering will be needed, together with improving the robustness of the SIMBA clustering.Acknowledgements
No acknowledgement found.References
1. Fogel, M. A. et al. Society for Cardiovascular Magnetic Resonance/European Society of Cardiovascular Imaging/American Society of Echocardiography/Society for Pediatric Radiology/North American Society for Cardiovascular Imaging Guidelines for the use of cardiovascular magnet. J. Cardiovasc. Magn. Reson. 24, 1–78 (2022).
2. Di Sopra, L., Piccini, D., Coppo, S., Stuber, M. & Yerly, J. An automated approach to fully self-gated free-running cardiac and respiratory motion-resolved 5D whole-heart MRI. Magn. Reson. Med. 82, 2118–2132 (2019).
3. Coppo, S. et al. Free-running 4D whole-heart self-navigated golden angle MRI: Initial results. Magn. Reson. Med. 74, 1306–1316 (2015).
4. Heerfordt, J. et al. Similarity-driven multi-dimensional binning algorithm (SIMBA) for free-running motion-suppressed whole-heart MRA. Magn. Reson. Med. 86, 213–229 (2021).
5. Piccini, D., Littmann, A., Nielles-Vallespin, S. & Zenge, M. O. Spiral phyllotaxis: The natural way to construct a 3D radial trajectory in MRI. Magn. Reson. Med. 66, 1049–1056 (2011).
6. Feng, L. et al. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn. Reson. Med. 75, 775–788 (2016).
7. Modat, M. et al. Fast free-form deformation using graphics processing units. Comput. Methods Programs Biomed. 98, 278–284 (2010).
8. Knoll, F., Schwarzl, A., Diwoky, C. & Sodickson, D. K. gpuNUFFT - An open source GPU library for 3D regridding with direct Matlab interface. Proc. Intl. Soc. Mag. Reson. Med. 4297 (2014).
9. Ahmad, R., Ding, Y. & Simonetti, O. P. Edge Sharpness Assessment by Parametric Modeling: Application to Magnetic Resonance Imaging. Concepts Magn. Reson. Part A. Bridg. Educ. Res. 44, 138–149 (2015).
10. Etienne, A. et al. ‘Soap-Bubble’ visualization and quantitative analysis of 3D coronary magnetic resonance angiograms. Magn. Reson. Med. 48, 658–666 (2002).
11. Piccini, D. et al. Deep Learning to Automate Reference-Free Image Quality Assessment of Whole-Heart MR Images. Radiol. Artif. Intell. 2, e190123–e190123 (2020).
Figures
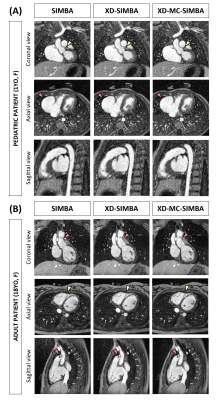
(A) 1-year-old female patient (also shown in Figure 2) with an aneurysm of the right atrial appendage. The image is zoomed around the heart (heart diameter <10cm).
(B) 29-year-old male patient with a bicuspid aortic valve and dilated aortic root.
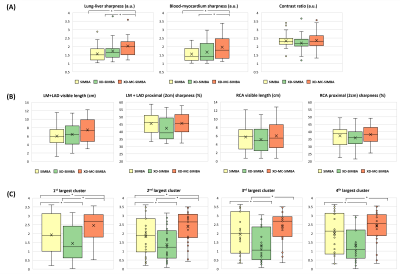
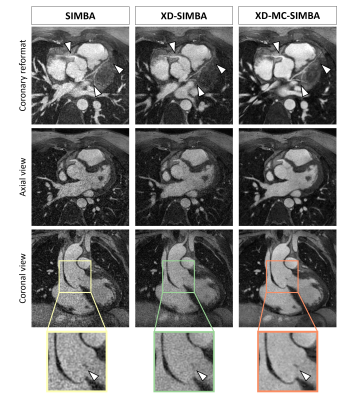
Figure 5. Example of a patient after repair of tetralogy of Fallot and zoomed-in view of the aortic valve. The coaptation area of the aortic leaflet (arrow) is more clearly visible in XD-MC-SIMBA. In this example, XD-MC-SIMBA has a much higher assigned image quality score, while XD-SIMBA led to only a slight improvement in image quality relative to SIMBA. When looking at the coronary reformat, the distal portions of the left anterior descending and left circumflex coronary arteries are only depicted in XD-MC-SIMBA, and the proximal right coronary artery is also better delineated.