1276
Unlocking 20-fold acceleration towards 0.5-second whole-brain HCP-style fMRI1Electrical and Computer Engineering, University of Minnesota, Minneapolis, MN, United States, 2Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States, 3Department of Neurosurgery, University of Minnesota, Minneapolis, MN, United States
Synopsis
Keywords: fMRI, fMRI
Functional MRI (fMRI) is acquired with simultaneous multi-slice (SMS) imaging and in-plane acceleration to provide sufficient coverage and spatio-temporal resolutions. However, further accelerations are desirable to achieve BRAIN initiative targets. In this work, we investigate self-supervised deep learning reconstruction at 20-fold (SMS×in-plane=5×4) retrospective and prospective accelerations. Results show DL at 20-fold retrospective acceleration is similar to split slice-GRAPPA at 10-fold acceleration. Furthermore, we show that DL method trained on retrospective 20-fold acceleration generalizes well and successfully reconstructs prospectively 20-fold accelerated fMRI data.INTRODUCTION
Functional MRI (fMRI) is typically acquired using simultaneous multi-slice (SMS) imaging1 in combination with in-plane acceleration for sufficient coverage and resolution. Yet higher acceleration rates are desired for improving spatio-temporal resolutions. Physics-guided deep learning (PG-DL) reconstruction has shown promise in highly-accelerated MRI, improving on conventional methods at higher accelerations2-6. Recently, a self-supervised PG-DL reconstruction, which does not require fully-sampled training data, was applied to HCP-style retinotopic mapping fMRI with an acceleration of 10-fold (SMS×in-plane =5×2), and was shown to improve tSNR while preserving retinotopic activation7,8. However, further studies are needed to establish its feasibility at higher accelerations9, especially with prospectively accelerated acquisitions. In this study, we study PG-DL at 20-fold (SMS×in-plane= 5×4) acceleration with a different visual block design paradigm, which enables characterizing a gold standard activation pattern. We first use retrospective acceleration for a head-to-head comparison of PG-DL with standard acquisitions. Then we show PG-DL trained with retrospective acceleration generalizes well to prospectively accelerated fMRI.METHODS
fMRI Data Acquisition:Imaging was performed on a 7T (Siemens Magnetom) with a 32-channel head coil in 6 subjects using HCP-style acquisitions: TR/TE/α=1s/21ms/70°; FOV=208×208mm2; spatial resolution=1.6×1.6×1.6mm3; SMS-factor=5; in-plane acceleration (R)= 2 and 1/3-FOV CAIPI shifts. 10 runs (~3mins/each) were acquired with a standard 24s on-off visual block design paradigm with a center/target and surrounded checkerboard counterphase flickering at 6 Hz. For retrospective acceleration, data were retrospectively undersampled in-plane to R=4 (20-fold total acceleration).
Additionally, one subject was re-imaged on a separate day using prospective 20-fold acceleration (SMS×R=5×4). Imaging parameters were kept the same to avoid any confounding factors that may arise from changes in TR/TE.
Self-Supervised PG-DL Reconstruction:
PG-DL reconstruction uses algorithm unrolling, alternating between data-fidelity (DF) and regularization5. Standard linear methods are used for the former, while the latter is implemented implicitly via neural networks. These were trained end-to-end over a database using self-supervised learning6,8 (SSDU), which splits acquired undersampled k-space into two disjoint sets, $$$\Theta$$$ and $$$\Lambda$$$. $$$\Theta$$$ enforces DF, while $$$\Lambda$$$ defines k-space loss6. Three sets of disjoint masks are used as data augmentation10. PG-DL network was implemented with 10 unrolls/iterations with ResNet8 as regularizers.
Training was performed on 4 subjects with retrospectively 20-fold accelerated scans (Learning-rate=10-4,mixed ℓ1-ℓ2 loss,100 epochs) Testing was performed on retrospective 20-fold acceleration (2 subjects unseen by the network) and on prospective 20-fold acceleration (1 unseen subject). Comparisons were made to split slice-GRAPPA (SP-SG)11.
Data Analysis:
Functional preprocessing included 3D rigid body motion correction and low-drift removal (3rd order DCT). GLM analyses were carried out on one run for each reconstruction independently. GLM design matrices were generated by convolving of a double gamma with a “box car” function, the latter representing the stimuli’s onsets and offsets.
RESULTS
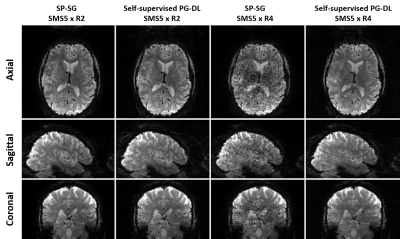
Retrospective acceleration:Fig. 1 shows representative reconstructed images. PG-DL reconstruction at 20-fold visibly reduces noise and aliasing/leakage observed in SP-SG, showing similar image quality to SP-SG at 10-fold acceleration.
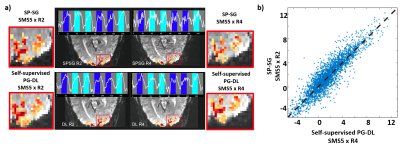
Fig. 2a depicts activations with t-values thresholded at t>3.6. Time courses belong to same single voxel within the red square. SP-SG at 20-fold does not preserve extent of activation and time courses, whereas PG-DL showed similar activation and time courses compared to standard acceleration. Single-voxel scatter plots (Fig.2b) for all voxels within a region of interest encompassing V1 for t-values for the contrast target and surround > 0 for SP-SG at 10-fold and PG- DL at 20-fold shows good agreement.
Prospective acceleration:
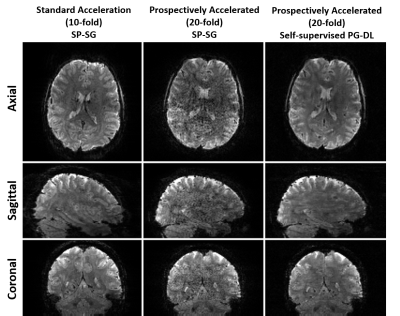
Fig. 3 shows representative reconstructed images acquired from the same subject in different scans. As with retrospective acceleration, PG-DL outperforms SP-SG at 20-fold, showing similar quality to SP-SG at 10-fold. This also establishes that PG-DL trained on retrospective acceleration generalizes to prospective acceleration.
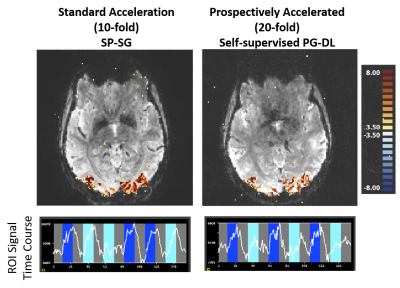
Fig. 4 shows the t-maps for the contrast target and surround > 0, and time courses for the combined activation of target and surround for the subject in Fig. 3. Time courses and t-maps look comparable even though scans were acquired on different days.
DISCUSSION AND CONCLUSIONS
In this study, we evaluated the feasibility of 20-fold acceleration on HCP-style 7T fMRI acquisitions. PG- DL reconstruction at 20-fold acceleration shows better image quality than SP-SG, and close to the SP-SG at standard 10-fold acceleration with highly-correlated activation patterns. We also showed, for the first time, that the PG-DL reconstruction trained on fMRI with 20-fold retrospective acceleration generalizes to 20-fold prospective acceleration. This is especially important since the higher in-plane acceleration affects the EPI train acquisition. Our paradigm exploited the retinotopic properties of early and primary visual cortices, which provide a ground truth with regards to the spatial extent of expected activation elicited by flickering grating stimuli. With the 20-fold prospective accelerated scans, not only can we observe the expected pattern of BOLD activation in relevant regions, but the t-values remain relatively large and close to those obtained from 10-fold images, indicating these reconstructions maintain the overall quality with precision.While for this study, we kept the sequence parameters (TR/TE/resolution) same for 20-fold prospective acceleration to establish feasibility, future studies will use this additional acceleration to push towards ~0.5s whole-brain TR and/or higher spatial resolutions. Further investigations using more runs, as well as acquiring prospectively accelerated data in the same session, are warranted.
Acknowledgements
Funding: Grant support: NIH, Grant numbers: P30 NS076408, R01 HL153146, U01 EB025144, P41 EB027061; NSF, Grant number: CAREER CCF-1651825.
∗: The first two authors contributed equally to this work.
References
[1] Moeller, Steen, et al. "Multiband multislice GE‐EPI at 7 Tesla, with 16‐fold acceleration using partial parallel imaging with application to high spatial and temporal whole‐brain fMRI." Magnetic Resonance in Medicine 63.5 (2010): 1144-1153.
[2] Knoll, Florian, et al. "Deep-learning methods for parallel magnetic resonance imaging reconstruction: A survey of the current approaches, trends, and issues." IEEE Signal Processing Magazine 37.1 (2020): 128-140.
[3] Hammernik, Kerstin, et al. "Learning a variational network for reconstruction of accelerated MRI data." Magnetic Resonance in Medicine 79.6 (2018): 3055-3071.
[4] Schlemper, Jo, et al. "A deep cascade of convolutional neural networks for dynamic MR image reconstruction." IEEE Transactions on Medical Imaging 37.2 (2017): 491-503.
[5] Aggarwal, Hemant K., Merry P. Mani, and Mathews Jacob. "MoDL: Model-based deep learning architecture for inverse problems." IEEE Transactions on Medical Imaging 38.2 (2018): 394-405.
[6] Yaman, Burhaneddin, et al. "Self‐supervised learning of physics‐guided reconstruction neural networks without fully sampled reference data." Magnetic Resonance in Medicine 84.6 (2020): 3172-3191.
[7] Demirel O B, Yaman B, Dowdle L, Moeller S, Vizioli L, Yacoub E, Strupp J, Olman C A, Ugurbil K and Akcakaya M 2021b Improved Simultaneous Multi-Slice Functional MRI Using Self-supervised Deep Learning. (IEEE: 55th Asilomar) pp 890-4.
[8] O. B. Demirel, B. Yaman, S. Moeller, L. Dowdle, L. Vizioli, K. Kay, E. Yacoub, J. Strupp, C. Olman, K. Ugurbil and M. Akçakaya."Improved Accelerated fMRI Reconstruction using Self-supervised Deep Learning" Proc. 29th Meeting of ISMRM, Virtual Conference, May 2021,
[9] Demirel, Omer Burak, et al. "20-fold Accelerated 7T fMRI Using Referenceless Self-Supervised Deep Learning Reconstruction." 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, 2021.
[10] Yaman, Burhaneddin, et al. "Multi‐mask self‐supervised learning for physics‐guided neural networks in highly accelerated magnetic resonance imaging." NMR in Biomedicine (2022): e4798.
[11] Cauley, Stephen F., et al. "Interslice leakage artifact reduction technique for simultaneous multislice acquisitions." Magnetic resonance in medicine 72.1 (2014): 93-102.
Figures