1225
3D MRI-US multimodal alignment for real-time intervention - a tradeoff between accuracy and computation time1GE Research, Niskayuna, NY, United States, 2Depts. of Radiology and Urology, University of Wisconsin-Madison, Madison, WI, United States, 3Dept. of Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 4Depts. of Radiology and Biomedical Engineering, University of Wisconsin-Madison, Madison, WI, United States, 5Dept. of Radiology, University of Iowa, Iowa City, IA, United States
Synopsis
Keywords: Liver, Multimodal, Real-time multimodal alignment, image-guided intervention, MRI-US deformable fusion
Multimodal MRI-US image fusion in image-guided therapy such as liver microwave ablation aids in correct placement of the applicator device. Significant tissue deformation due to breathing motion requires alignment of pre-interventional MRI on real-time interventional US to compensate for the motion. In this work, we present a hybrid framework (conventional and deep learning) for multimodal deformable registration to provide higher accuracy and retain the low latency required for real-time interventions.INTRODUCTION
Multimodal MRI-US fusion in image-guided therapy can enable greater precision and accuracy in targeting pathological soft tissues (more conspicuous with MRI) in conjunction with ultrasound (US) for real-time interventional guidance. In the case of needle-based microwave ablation (MWA) therapy in the liver, the placement of a MWA applicator at the tumor target under US guidance is a challenging task without pre-intervention MRI (pre-MRI) or CT due to poor lesion conspicuity in US. However, the liver deforms significantly when a subject breathes, which necessitates the alignment of pre-MRI to every interventional US (iUS) frame to compensate for motion. The alignment method needs to be fast (~3 fps) and accurate (<3mm). In this work, we present a hybrid approach, combining conventional and deep learning (DL) image registration that aligns pre-MRI with iUS in near real time with the desired accuracy. Compared to pure DL or conventional deformable registration approaches, this is a feasible solution that balances the dual-requirement for accurate and fast MRI-US registration for intervention guidance.METHOD
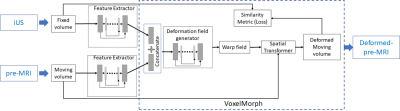
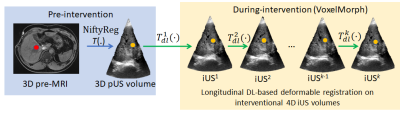
We leveraged the simultaneous MRI-US imaging platform1 with a handsfree MRI-compatible US probe2 that acquired pre-MRI with pre-interventional 3D US (pUS), and the same probe was used to acquire 3D iUS at ~3 volumes/s. For the multimodal alignment, we developed/used the following: A) a conventional multimodal deformable registration (R-TRAD) as a baseline using NiftyReg3, which is a combination of affine registration based on image-patch similarity (cross-correlation), and B-Spline for deformable registration, B) a DL-based architecture (Fig. 1) that has UNet4 in feature extraction part for MRI and US separately before using DL-based VoxelMorph5 (a deformable registration method involving UNet and spatial transformation with high computational efficiency) for direct pre-MRI to pUS/iUS alignment (R-DL), C) a hybrid framework (Fig. 2) that uses NiftyReg for pre-MRI to pUS alignment and VoxelMorph for pUS-iUS alignment (R-HYBRID). Since pre-MRI-pUS alignment is required only during treatment planning (pre-intervention), it is not time-critical and thus the longer computation time of conventional deformable registration can be tolerated. However, real-time computation of deformation fields between pUS-iUS is crucial for adapting to the deformation across longitudinal iUS volumes. The deformation fields from NiftyReg and DL registration were composed together to obtain a pre-MRI-iUS alignment in the hybrid framework. Normalized cross correlation (NCC) was used as a loss function, and 100/500 epochs with learning rates of 1e-4 & 1e-5 were used to train the methods in (B) & (C) respectively.We acquired respiratory-gated T1w-MRI (pre-MRI) & US volumes of the liver from 3 volunteers with informed consent and IRB approval. The pre-MRI and ~1600 US volumes from one volunteer were used to train the DL networks, and testing was done with pre-MRI and 20 longitudinal pUS/iUS volumes across all volunteers (excluding the iUS volumes used for training). A set of 2-3 expert-placed landmarks were used to report the landmark distances (mean Euclidean distances) before and after multimodal alignment.
RESULTS
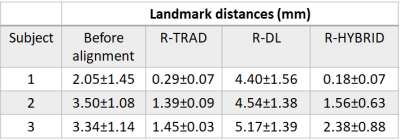
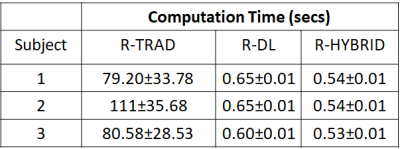
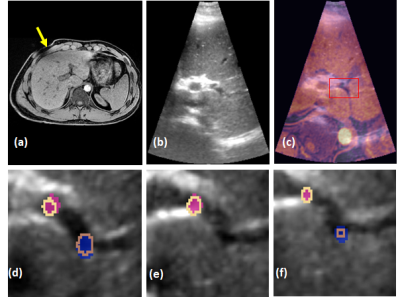
Table 1 shows the mean landmark distances between the pre-MRI-iUS before and after deformable alignment using the R-TRAD, R-DL and R-HYBRID methods for the 3 volunteers across 20 volume pair registrations. The results show that both R-TRAD and R-HYBRID had reduced landmark distances after registration, while R-DL provided higher errors in landmarks. Table 2 shows the average computation times for R-TRAD, R-DL and R-HYBRID (excluding the conventional registration times). Fig. 3 shows the qualitative results of the R-HYBRID method demonstrating precise alignment of the deformed pre-MRI and pUS/iUS landmarks for one subject.DISCUSSIONS AND CONCLUSIONS
This is a preliminary study comparing the performances of MRI-US deformable registration methods in liver and identifying the ones that may be potentially used for real time intervention. Although conventional deformable registration provided the highest accuracy in terms of landmark distances, the computation times were unsuitable for real time intervention. The failure of the R-DL method can be attributed to lack of paired multimodal data and enough variation, and that DL methods are probably better suited for single modality alignment. This observation is consistent with the existing literature6,7, where the best performing image alignment method for MRI-US datasets involved a conventional unsupervised method and not a learning approach. R-HYBRID allowed us to achieve a tradeoff between the accuracy of alignment and computation times required for real time image-guided interventions. We believe such hybrid methods will be critical to the success of leveraging multimodal fusion in interventional environment.Acknowledgements
This research was supported by NIH/NCI grant number 1R01CA266879.References
1) Bednarz B, Jupitz S, Lee W, et al. First-in-human imaging using a MR-compatible e4D ultrasound probe for motion management of radiotherapy. Phys Med. 2021; 88:104-110.
2) Lee W, Chan H, Chan P, et al. A magnetic resonance compatible E4D ultrasound probe for motion management of radiation therapy. IEEE network. 2017; 017:10.1109/ULTSYM.2017.8092, 223.
3) Modat M, Ridgway GR, Taylor ZA, et al. Fast free-form deformation using graphics processing units. Computer methods and programs in biomedicine. 2010; 98(3):278-284.
4) Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Proc of the Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer. 2015; LNCS, 9351: 23-241.
5) Balakrishnan G, Zhao A, Sabuncu MR, et al. Voxelmorph: A learning framework for deformable medical image registration. IEEE Trans on Medical Imaging. 2019; 38(8):1788-1800.
6) Xiao Y, Rivaz H, Chabanas M, et al. Evaluation of MRI to ultrasound registration methods for brain shift correction: The CuRIOUS2018 Challenge. IEEE Trans on Medical Imaging. 2019; 39(3):777-786.
7) Hering A, Hansen L, Mok TCW, et al. Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning. IEEE Trans on Medical Imaging. 2022; doi: 10.1109/TMI.2022.3213983.
Figures