1217
Improved Free-Breathing Volumetric Liver Fat and R2* Quantification Using 3D Stack-of-Radial GRE Dixon MRI and XD-GRASP Reconstruction
Xiaodong Zhong1, Marcel D Nickel2, Stephan A.R. Kannengiesser2, Brian M Dale3, Holden H Wu4, and Vibhas Deshpande5
1MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Los Angeles, CA, United States, 2MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany, 3MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Cary, NC, United States, 4Department of Radiological Sciences, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, United States, 5MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Austin, TX, United States
1MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Los Angeles, CA, United States, 2MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany, 3MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Cary, NC, United States, 4Department of Radiological Sciences, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, CA, United States, 5MR R&D Collaborations, Siemens Medical Solutions USA, Inc., Austin, TX, United States
Synopsis
Keywords: Liver, Quantitative Imaging, Fat, R2*, Iron
Respiratory motion compensation is necessary for free-breathing stack-of-radial liver fat and R2* quantification. While self-gating is a valid approach, it may lead to degraded quality of images and quantitative maps and possible prolonged acquisition. A 3D XD-GRASP stack-of-radial technique was developed and evaluated in a motion phantom and in vivo subjects. Results demonstrated PDFF and R2* agreement of the proposed method compared to reference methods. Improved image and map quality and PDFF and R2* quantification agreement of the proposed method using an acceleration factor of 4 (equivalent to 105 seconds of time saving) were observed compared to the self-gating method.INTRODUCTION
For liver proton density fat fraction (PDFF) and R2* quantification, breath-hold 3D multi-echo Cartesian GRE MRI is clinically used1-3. 3D stack-of-radial imaging is promising for accurate free-breathing liver PDFF and R2* quantification4-8. While self-gating is necessary to compensate the respiratory motion influence on free-breathing stack-of-radial R2* quantification, it may lead to residual radial undersampling artifacts and possible prolonged acquisition6-8.Stack-of-radial MRI with extra-dimensional golden-angle radial sparse parallel (XD-GRASP) reconstruction has demonstrated improved image quality in applications that use undersampled radial data such as dynamic contrast-enhanced liver imaging9 and liver T1 mapping10. The purpose of this study was to develop an XD-GRASP stack-of-radial MRI technique for PDFF and R2* quantification and validate its performance in a motion phantom and in vivo subjects.
METHODS
XD-GRASP ReconstructionWith the motion-state dimension resolved by self-gating being the extra dimension, undersampled stack-of-radial data was reconstructed by optimization of a cost function ½*||Ax-y||22+||Wx||1, where A is a catch-all matrix collectively applying all relevant operations, x is the image matrix to reconstruct, y is the multi-channel k-space data, W is the redundant Haar wavelet transformation applied in spatial and motion-state dimensions, ||·||1 is the l1 norm. W can be configured to regularize on spatial-only constraints, or on both spatial and temporal (motion-state) constraints.
Pulse Sequences and Image Reconstruction
All radial data were acquired using a multi-echo stack-of-radial research application sequence6,7. Several techniques were used for reconstruction. Self-gating was applied to accept data acquired near end-expiration with a 40% acceptance rate as the current method6,7. Alternately, self-gating was used to resolve 4 respiratory motion states. Data of the end-expiration motion state was regularized by spatial-only and spatial-temporal constraints (the proposed method, XD-GRASP) respectively with empirical configurations.
Phantom Data and Analysis
From a previous study7, stack-of-radial MRI data of a custom phantom scanned at 3T (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany) was processed. The phantom had 4 vials containing ferumoxytol solutions (Feraheme, AMAG Pharmaceuticals, Waltham, MA, USA) with concentrations of 0, 25, 37 and 50 ug/mL, a saline bag and a plastic holder filled with agar gel. During the acquisitions, a motion stage was used to move the holder and 4 vials according to a pre-recorded respiratory motion waveform. Imaging parameters are listed in Table 1. The acquisition was repeated with the phantom being stationary.
Raw data were undersampled to 128 views, equivalent to an acceleration factor of 4 (based on 512 regarded as fully sampled). Mono-exponential fitting was used to calculate R2*. Regions of interest (ROIs) were placed inside the vials, holder and saline bag. R2* values were compared to reference values without motion using Bland-Altman analysis with mean difference (MD) and limits of agreement (LoA=MD±1.96×SD).
In Vivo Data and Analysis
This study was HIPAA-compliant and approved by the local IRB. Data included in a previous work6 were analyzed: free-breathing stack-of-radial and breath-hold Cartesian data from 5 healthy subjects (1 female, 34.6±4.8 yrs, body mass index [BMI]: 22.6±3.5 kg/m2) and 6 subjects with non-alcoholic fatty liver disease (3 females, 58.5±9.5 yrs, BMI: 28.9±2.7 kg/m2) acquired at 3T (MAGNETOM Prisma or Skyra, Siemens Healthcare, Erlangen, Germany). Imaging parameters are listed in Table 1.
Raw data were undersampled by a factor of 4 for comparison. Multi-step adaptive fitting was performed for PDFF and R2* quantification3. Twelve ROIs were placed on four slices in the maps. PDFF and R2* values were compared to reference breath-hold Cartesian values using Bland-Altman plots with standard deviation values being error bars. The R2* values were also fit to a linear mixed effects model with the methods and number of views as interacting fixed effects and the subject as a random effect.
RESULTS
Example images, R2* maps and Bland-Altman plots of the phantom using data with 128 views are shown in Figure 1. Improved quality of echo images and R2* maps was evident by the proposed XD-GRASP method compared to the current self-gating method.Example images and R2* maps of one subject using data with 404 and 101 views are shown in Figure 2 and 3. Improved quality of images and maps using XD-GRASP was also observed, especially for 101 views. In Figure 4, Bland-Altman plots showed improved MD, LoA and error bars of the proposed method compared to the current method for 101 views.
No method had significantly different (p>0.998) mean of R2* values compared to breath-hold Cartesian data. XD-GRASP had similar (p>0.966) SD of R2* values compared to breath-hold Cartesian, and significantly lower (p<0.022) SD of R2* values compared to all other methods except for the average method with 404 views (p=0.999).
DISCUSSION AND CONCLUSION
A 3D stack-of-radial GRE Dixon MRI technique was developed for free-breathing liver PDFF and R2* quantification and evaluated in a motion phantom and in vivo subjects. Results demonstrated agreement of PDFF and R2* measured by the proposed method compared to the reference methods. Improved image and map quality and PDFF and R2* quantification agreement of the XD-GRASP method were observed compared to the current self-gating method with an acceleration factor of 4, equivalent to an approximate acquisition time saving of 105 seconds for the in vivo protocols in this study. This proposed method may allow more efficient free-breathing liver PDFF/R2* mapping.Acknowledgements
This study was supported in part by Siemens Medical Solutions USA, Inc. and the Department of Radiological Sciences at UCLA.References
1. Yu H, Shimakawa A, McKenzie CA, Brodsky E, Brittain JH, Reeder SB. Multiecho water‐fat separation and simultaneous R2* estimation with multifrequency fat spectrum modeling. Magn Reson Med 2008;60:1122-1134.2. Hernando D, Levin YS, Sirlin CB, Reeder SB. Quantification of liver iron with MRI: state of the art and remaining challenges. J Magn Reson Imaging 2014;40:1003-1021.
3. Zhong X, Nickel MD, Kannengiesser SAR, Dale BM, Kiefer B, Bashir MR. Liver fat quantification using a multi‐step adaptive fitting approach with multi‐echo GRE imaging. Magn Reson Med 2014;72:1353-1365.
4. Armstrong T, Dregely I, Stemmer A, et al. Free-breathing liver fat quantification using a multiecho 3D stack-of-radial technique. Magn Reson Med 2018;79:370-382.
5. Armstrong T, Ly KV, Murthy S, et al. Free-breathing quantification of hepatic fat in healthy children and children with nonalcoholic fatty liver disease using a multi-echo 3-D stack-of-radial MRI technique. Pediatr Radiol 2018;48:941-953.
6. Zhong X, Armstrong T, Nickel MD, Kannengiesser SAR, Pan L, Dale BM, Deshpande V, Kiefer B, Wu HH. Effect of respiratory motion on free-breathing 3D stack-of-radial liver R2* relaxometry and improved quantification accuracy using self-gating. Magn Reson Med 2020;83:1964-1978.
7. Zhong X, Hu HH, Armstrong T, Li X, Lee YH, Tsao TC, Nickel MD, Kannengiesser SAR, Dale BM, Deshpande V, Kiefer B, Wu HH. Free-breathing volumetric liver R2* and proton density fat fraction quantification in pediatric patients using stack-of-radial MRI with self-gating motion compensation. J Magn Reson Imaging 2021;53:118-129.
8. Armstrong T, Zhong X, Shih SF, Felker E, Lu DS, Dale BM, Wu HH. Free-breathing 3D stack-of-radial MRI quantification of liver fat and R2* in adults with fatty liver disease. Magn Reson Imaging 2022;85:141-152.
9. Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med 2016;75:775-788.
10. Feng L, Liu F, Soultanidis G, Liu C, Benkert T, Block KT, Fayad ZA, Yang Y. Magnetization-prepared GRASP MRI for rapid 3D T1 mapping and fat/water-separated T1 mapping. Magn Reson Med 2021;86:97-114.
11. Breuer FA, Blaimer M, Mueller MF, Seiberlich N, Heidemann RM, Griswold MA, Jakob PM. Controlled aliasing in volumetric parallel imaging (2D CAIPIRINHA). Magn Reson Med 2006;55:549-556.
Figures
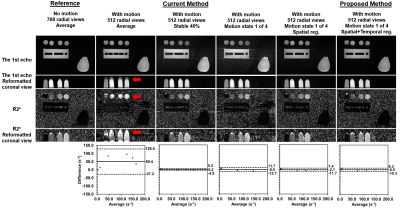
Figure 1
Example images, R2* maps and Bland-Altman plots of the phantom using
data with 128 radial views. The ground truth is the results from the data with
768 radial views acquired when the phantom was not moving (the 1st column). All
the other methods show results from the data with 128 radial views acquired
when the phantom was moving (the 2nd to 6th columns). All Bland-Altman plots
show comparison between the corresponding method and the ground truth. Red
arrows indicate the motion-induced artifacts in the images and maps.
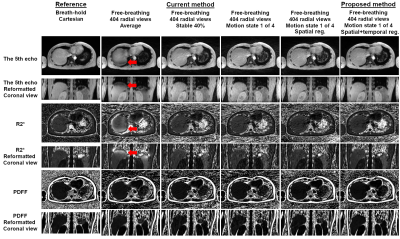
Figure 2
Example images and R2* maps
of one subject reconstructed using data with 404 radial views. The reference results
are from the breath-hold Cartesian acquisitions (the 1st column). All the other
methods show results from the data with 404 radial views (the 2nd to 6th columns).
Red arrows indicate the motion-induced artifacts in the images and maps.
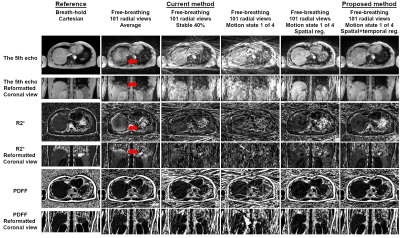
Figure 3
Example images and R2* maps
of one subject reconstructed using data with 101 radial views (acceleration
factor = 4). The reference results are from the breath-hold Cartesian
acquisitions (the 1st column). All the other methods show results from the data
with 404 radial views (the 2nd to 6th columns). Red arrows indicate the
motion-induced artifacts in the images and maps.
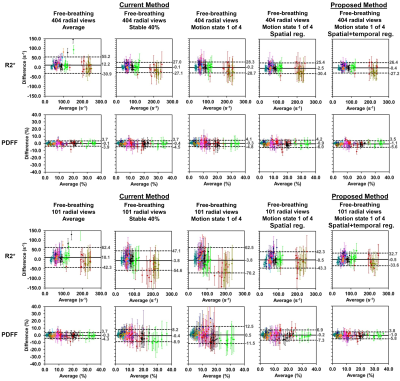
Figure 4
Bland-Altman plots comparing the R2* and PDFF results from different methods vs. those from the reference
breath-hold Cartesian acquisitions for data with 404 and 101 radial views
respectively. The difference was obtained by results from free-breathing
stack-of-radial acquisitions subtracted results from the reference breath-hold
Cartesian acquisitions. Different colors represent the values within 12 ROIs of
different subjects. The error bars are the standard deviation of measured R2*
or PDFF values within the ROI of each method.
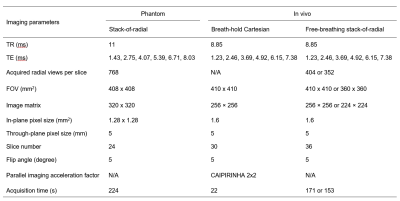
Table
1 Imaging parameters of the protocols
used for the phantom and in vivo data acquisition. N/A: Not applicable.
CAIPIRINHA: Two-dimensional parallel imaging as described in11.
DOI: https://doi.org/10.58530/2023/1217