1171
Unsupervised learning for MRI cross-scanner harmonization1Auckland Bioengineering Institue, university of Auckland, Auckland, New Zealand, 2Mātai Medical Research Institute, Gisborne, New Zealand, 3Department of Computer Science, Faculty of Science, University of Auckland, Auckland, New Zealand, 4Department of Anatomy and Medical Imaging, Faculty of Medical and Health Sciences, University of Auckland, Auckland, New Zealand
Synopsis
Keywords: Multi-Contrast, Machine Learning/Artificial Intelligence, harmonization, normalization, reconstruction
Harmonization is necessary for large-scale multi-site neuroimaging studies to reduce the variations due to factors such as image acquisition, imaging devices, and acquisition protocols. This so-called scanner effect significantly impacts multivariate analysis and the development of computational predictive models using MRI. Our approach utilized an unsupervised learning based model to build a mapping between MR data acquired from two different scanners. Results illustrate the potential of unsupervised deep learning algorithms to harmonize MRI data, as well as to improve downstream tasks by applying the harmonization.Introduction
Pooling data across multiple centres can increase the statistical power of neuroimaging research. However, combining datasets from different research centres is challenging due to differences in image acquisition, demographics, imaging devices, acquisition protocols, image quality, and other factors1,2. To minimize these non-biological variations, we propose an unsupervised learning approach based on generative adversarial networks (GAN)3. GAN is a deep learning model that learns a mapping between two domains while preserving the original semantic information in the transformed data by ensuring that a mapping back to the original domain is enforced. This study investigated the feasibility of using a GAN model to harmonize volumetric data obtained from T1-weighted MR images.Methods:
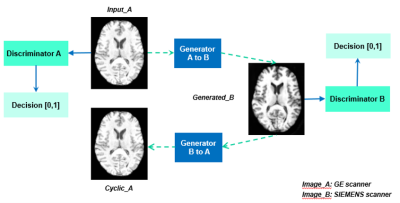
A public dataset of 104 3T scans was acquired from GE and SIEMENS MRI scanners were selected from Parkinson's Progression Markers Initiative (PPMI)4 database and SRPBS5 Traveling Subject MRI Dataset. These images formed the input and reference domains. Unpaired scans from the PPMI dataset were used to train the model using data from the SIEMENS scanner as the reference. CycleGAN is a dual generator-discriminator network (Figure 1), and we adapted CycleGAN and used a U-net generator with L1 loss and patchGAN for the discriminator.A direct evaluation was performed on the paired scans using image properties metrics, including the mean squared error (MSE), structural similarity index metric (SSIM), the peak signal-to-noise ratio (PSNR), and histogram correlation (HC). PCA site clustering, along with Support Vector Machine (SVM)6 classifiers for scanner classification, was applied to radiomics features and was used to examine if the scanner effect had been reduced. An SVM classifier to distinguish Parkinson's disease patients and normal controls was trained following a five fold-cross-validation procedure to investigate whether machine learning predictor can be improved by applying harmonization.
Results:
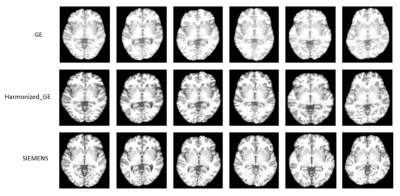
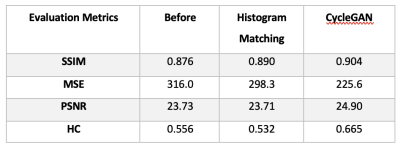
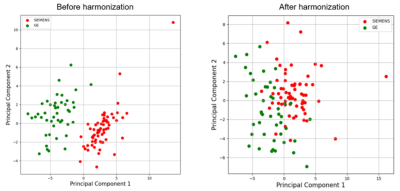
Contrast similarity between the harmonized (GE to SIEMENS) and SIEMENS datasets could be visually inspected (Figure 2). Four performance indicators (MSE, SSMI, PSNR and HC) were improved with the proposed method (Figure 3). Compared to the raw image, the PSNR, HC, and SSIM were increased by 4.9%, 19.6%, and 0.3%, respectively, while the MSE was reduced by 28.6% after applying harmonization. Furthermore, our approach outperformed the histogram matching method.Figure 4 illustrates the harmonization effects on site classification. After applying the harmonization, the two site clusters were getting closer to each other, and the accuracy of the SVM site prediction decreased from 0.95% to 0.59% (before and after harmonization, respectively). Regarding the classification of Parkinson's disease patients, results showed that after applying harmonization, the accuracy and AUC were improved. Accuracy was increased from 0.55% to 0.63%, and the AUC was increased from 0.54 to 0.59.
Discussion:
This work demonstrates the effectiveness of 3D CycleGAN applied to the harmonization of the T1-weighted MR images. Results showed that the MSE, SSIM, PSNR, HC, and classifier performance all improved after harmonization. The results of our experiments indicate that MRI data with a variety of different settings can be efficiently harmonized, which can be helpful in downstream neuroimaging analyses.The feasibility of using CycleGAN for harmonization has been demonstrated. However, more rigorous validation techniques are required to ensure that the harmonized image contains all of the original biological information while no new information is added since some studies noticed that GAN-based models might create new quantitative information7,8. Therefore, developing a robust method to evaluate and visualize the effect of harmonization is necessary. Moreover, the current model can only map two scanners; future work should extend this model to train more domains simultaneously.
Conclusion:
In this work, we utilized a CycleGAN to harmonize volumetric data on 3D spaces. Without the need for traveling subjects or any paired scanner or modalities of images from the same subjects, the model can ensure realistic outputs and allows the removal of cross-scanner bias while preserving the biological features. As part of this study, a large-scale analysis will be performed in the future to allow images from various domains to be harmonised.Acknowledgements
This work was partially supported by the Ministry of Business, Innovation and Employment (MBIE) of New Zealand [grant number PROP70240-CNZSDS-UOA] and the Health Research Council of New Zealand [grant number 21/144].References
1. Jovicich, J. et al. Multisite longitudinal reliability of tract-based spatial statistics in diffusion tensor imaging of healthy elderly subjects. NeuroImage 101, 390–403 (2014).
2. Fox, R. J. et al. A validation study of multicenter diffusion tensor imaging: reliability of fractional anisotropy and diffusivity values. AJNR Am. J. Neuroradiol. 33, 695–700 (2012).
3. Zhu, J.-Y., Park, T., Isola, P. & Efros, A. A. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. in 2017 IEEE International Conference on Computer Vision (ICCV) 2242–2251 (2017). doi:10.1109/ICCV.2017.244.
4. Parkinson Progression Marker Initiative. The Parkinson Progression Marker Initiative (PPMI). Prog. Neurobiol. 95, 629–635 (2011).
5. Tanaka, S. C. et al. A multi-site, multi-disorder resting-state magnetic resonance image database. Sci. Data 8, 227 (2021).
6. Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
7. Conditional Generative Adversarial Networks (cGANs) aided motion correction of dynamic 18F-FDG PET brain studies | Journal of Nuclear Medicine. https://jnm.snmjournals.org/content/early/2020/11/27/jnumed.120.248856.
8. Shiyam Sundar, L. K., Muzik, O., Buvat, I., Bidaut, L. & Beyer, T. Potentials and caveats of AI in hybrid imaging. Methods 188, 4–19 (2021).
Figures