1146
IVIM reconstruction from highly under-sampled DW-PROPELLER acquisition data via synthetic data-driven physics-informed deep learning1Department of Electronic Science, Fujian Provincial Key Laboratory of Plasma and Magnetic Resonance, Xiamen University, Xiamen, China, 2Department of Radiology, Zhongshan Hospital of Xiamen University, School of Medicine, Xiamen University, Xiamen, China, 3Clinical & Technical Solutions, Philips Healthcare, Shenzhen, China
Synopsis
Keywords: Quantitative Imaging, Diffusion/other diffusion imaging techniques
A synthetic data-driven physics-informed network (SDDPI-Net) was proposed for intravoxel incoherent motion (IVIM) mapping based on highly under-sampled diffusion-weighted turbo spin echo PROPELLER (DW-TSE-PROPELLER) data. This reconstruction network directly estimated distortion-free and artifacts-free IVIM parameters by explored data redundancy in the k-b space and IVIM bi-exponential model with synthetic training data. The results of human brain experiments show that our method can significantly improve the accuracy of IVIM maps with 6´ under-sampled DW-TSE-PROPELLER than other methods.Introduction
Fully-sampled DW-TSE-PROPELLER is more robust to geometric distortion and motion artifacts than diffusion-weighted single-shot / multi-shot echo planar imaging (DW-ssEPI or DW-msEPI), but it requires longer acquisition time for IVIM mapping.1,2 An advantage of DW-TSE-PROPELLER is the data redundancy in the k-space center, which makes it possible to accelerate data acquisition by under-sampling the k-space of each DW image with fewer blades.3 However, the effect of under-sampled artifacts during IVIM parameter fitting needs to be investigated. Therefore, we rotated blades along the b-value dimension to cover sufficient k-space information and developed a synthetic data-driven physics-informed network (SDDPI-Net) for rapid IVIM fitting. Compared with k-t SLR4 and subspace-LLR5, SDDPI-Net could directly estimate more accurate parameter maps without an extra voxel-wise fitting method by an end-to-end deep learning framework. Meanwhile, SDDPI-Net provides better-preserved tissue boundaries than other physical-informed deep learning methods DEEPIVIM6, IVIMNET7, and MANTIS8.Methods
Pulse sequence, data preprocessing, and synthetic data generation: Figure 1(a) shows the sequence diagram of DW-TSE-PROPELLER acquisition. Figure 1(b) shows the twelve-blade and two-blade schemes used for fully-sampling and under-sampling the k-space of each DW image, respectively. The data preprocessing pipeline is shown in Figure 1(c). Figure 1(d) shows the procedure of generating input-label pairs (x0, x) as training and validation data sets from realistic IVIM maps. According to the IVIM bi-exponential model, the signal intensity at a specific b-value, Sb, is defined by the following equation:$$S_b(S_0,D,f,D^*)=S_0 \times [f \times e^{-b \times D^*}+(1-f) \times e^{-b \times D}]$$The input-label pairs were split into 3902/879 pairs for network training/validation.
Network: SDDPI-Net is based on DIDN (Deep Iterative Down-Up) network9. DIDN was used as de-aliasing network g(·;θ1) to learn artifacts-free information from DW-TSE-PROPELLER images x0. The parameter estimate network h(·;θ2) consists of two fully connected layers with PReLU (parametric rectified linear unit). These fully connected layers can effectively map from learned information to four features m (m1, m2, m3, m4). Three modified sigmoid functions modulate feature values (m2, m3, m4) to physically plausible IVIM parameters (D, D*, and f). The modulation scalars of D, D*, and f lay in [0, 3000] μm2/s, [0, 50000] μm2/s, and [0, 1.0], respectively. $$\begin{cases} S_0=m_1 \\ D=D_{min}+sigmoid(m_2)\times (D_{min}-D_{max}) \\ D^*=D_{min}^*+sigmoid(m_3)\times (D_{min}^*-D_{max}^*) \\ f=f_{min}+sigmoid(m_4)\times (f_{min}-f_{max}) \end{cases}$$ The estimated physically plausible values for S0, D, f, and D* were substituted into the IVIM bi-exponential model to generate artifacts-free DW-TSE-PROPELLER images xrec.
An l1-norm between the predicted signal xrec from SDDPI-Net and fully-sampled DW-TSE-PROPELLER images x was used as a loss term. The overall architecture of SDDPI-Net is shown in Figure 3.
Experiments: In vivo data were acquired from two healthy volunteers on a Philips Ingenia CX 3.0T MRI scanner with a 16-channel head coil. Participants were instructed for four scans: (1) DW-ssEPI scan, (2) DW-msEPI scan, (3) fully-sampled DW-TSE-PROPELLER scan, and (4) under-sampled DW-TSE-PROPELLER scan. All scans were acquired with FOV = 220 mm × 220 mm, repetition time (TR) = 3000 ms, slice number = 8, slice thickness = 4 mm, 12 b-values. The other parameters for under-sampling DW-TSE-PROPELLER included: echo time (TE) = 106 ms, echo train length (ETL) per blade = 16, 2 blades per b-value, an effective matrix diameter of 128. DW-ssEPI (other parameters: TE = 145 ms, ETL = 128) / DW-msEPI (other parameters: TE = 69 ms, ETL = 32, 4 shots per b-value) and fully-sampling DW-TSE-PROPELLER (TE = 106 ms, 12 shots per b-value) scans were used as references to compare distortion resistance and quantitative accuracy, respectively.
Results
Figure 3 compares the zoomed-in results of DW-TSE-PROPELLER and EPI sequences in the forebrain region susceptible to geometric distortion. The anti-distortion ability of the DW-TSE-PROPELLER is based on the characteristic of TSE sequence. Therefore, the results of DW-TSE-PROPELLER were consistent with the reference TSE image. Whereas the results of DW-ssEPI and DW-msEPI had obvious geometric distortion. SDDPI-Net produced distortion-free IVIM parametric maps from the data acquired within similar acquisition time to DW-msEPI.Figure 4 shows IVIM parametric maps of an in vivo human brain estimated from different reconstruction methods. Incoherent artifacts caused by inconsistent trajectory per b-value corrupted the structural information in the zero-filled fitted D and f maps. Subtle missing details and inhomogeneous appearance still exist in other methods. The results of SDDPI-Net have sharper contrast and homogeneous appearance.
Table 1 summarizes the mean normalized root mean square error (nRMSE) and structural similarity index(SSIM) between the reference maps and the estimated maps over all 16 testing slices. SDDPI-Net yielded the small reconstruction errors and the highest similarity to the reference at R = 6.
Discussion and conclusion
SDDPI-Net can provide distortion-free and artifacts-free IVIM parametric maps from under-sampled DW-TSE-PROPELLER data. It solves under-sampling reconstruction and IVIM parameter fitting in an end-to-end framework, so it outperforms other combination methods. Compared with MANTIS, the robustness of the reconstruction of SDDPI-Net is achieved by constraining the output of SDDPI-Net to physically plausible IVIM parameter ranges via modifying sigmoid function.Acknowledgements
This work was supported in part by the Nation Natural Science Foundation of China under 11775184, 82071913, U1805261, and 22161142024.References
1. Jeon JY, Chung HW, Lee MH, et al. Usefulness of diffusion-weighted MR imaging for differentiating between benign and malignant superficial soft tissue tumours and tumour-like lesions. Br J Radiol. 2016;89:20150929.
2. Attenberger UI, Runge VM, Stemmer A, et al. Diffusion weighted imaging: a comprehensive evaluation of a fast spin echo DWI sequence with BLADE (PROPELLER) k-space sampling at 3 T, using a 32-channel head coil in acute brain ischemia. Invest Radiol. 2009;44:656-661.
3. Arfanakis K, Tamhane AA, Pipe JG, Anastasio MA. k-space undersampling in PROPELLER imaging. Magn Reson Med. 2005;53:675-683.
4. Lingala SG, Hu Y, DiBella E, et al. Accelerated dynamic MRI exploiting sparsity and low-rank structure: k-t SLR. IEEE Trans Med Imaging. 2011;30:1042-1054.
5. Wen Q, Feng L, Zhou K, et al. Rapid golden-angle diffusion-weighted propeller MRI for simultaneous assessment of ADC and IVIM. Neuro. Image. 2020; 223(1):117327.
6. Barbieri S, Gurney-Champion OJ, Klaassen R, et al. Deep learning how to fit an intravoxel incoherent motion model to diffusion-weighted MRI. Magn Reson Med. 2020;83:312-321.
7. Kaandorp MPT, Barbieri S, Klaassen R, et al. Improved unsupervised physics-informed deep learning for intravoxel incoherent motion modeling and evaluation in pancreatic cancer patients. Magn Reson Med. 2021;86:2250-2265.
8. Liu F, Feng L, Kijowski R. MANTIS: model-augmented neural network with Incoherent k-space sampling for efficient MR parameter mapping. Magn Reson Med. 2019;82:174-188.
9. Yu S, Park B, Jeong. J. Deep iterative down-up CNN for image denoising. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Long Beach, USA. 2019:2095-2103.
Figures
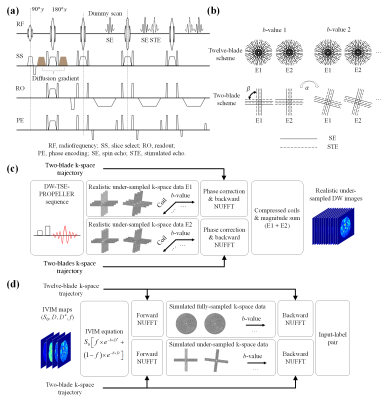
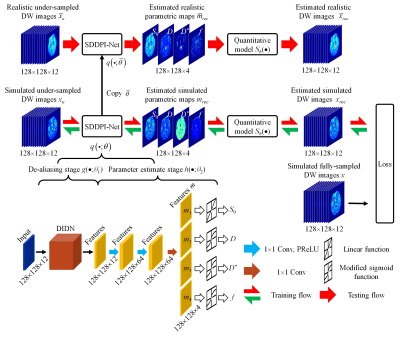
Figure 2. Overall architecture of the proposed SDDPI-Net for DW-TSE-PROPELLER MR image reconstruction. Under-sampled DW images as the input of SDDPI-Net are acquired at 12 b-values with the DW-TSE-PROPELLER sequence.
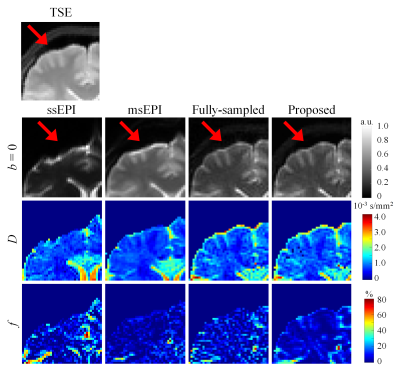
Figure 3. Distortion comparison between DW-TSE-PROPELLER imaging and EPI. A noticeable distortion (red arrow) lies in b = 0 DW-ssEPI and DW-msEPI images. Compared with DW-ssEPI or DW-msEPI, the results of TSE-based sequences (i.e., DW-TSE-PROPELLER or TSE) have no distortion.
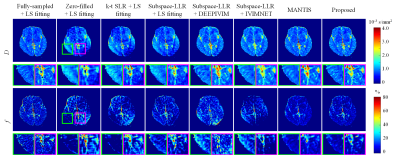
Figure 4. Representative axial slice DW images and IVIM parametric maps of an in vivo human brain estimated from different reconstruction + fitting methods. The fitted D and f maps are shown in the first to second rows and the zoomed-in areas are shown at the bottom of the corresponding maps. (LS, least-square)
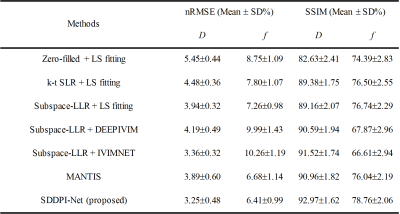
Table 1. Normalized RMSE and SSIM between the fully-sampled references and the estimated D and f maps on the in vivo human brain data set.