1105
Zero-DeepSub: Zero-Shot Deep Subspace Reconstruction for Multiparametric Quantitative MRI Using QALAS1Athinoula A. Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Massachusetts Institute of Technology, Cambridge, MA, United States, 4Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 5Fetal-Neonatal Neuroimaging & Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 6Harvard/MIT Health Sciences and Technology, Cambridge, MA, United States
Synopsis
Keywords: Quantitative Imaging, Quantitative Imaging
The 3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) has been developed and used for acquiring high-resolution T1, T2, and PD maps from five measurements within each repetition time. However, it assumes that each k-space data is acquired instantly at the first echo train length index neglecting T1 and T2 relaxation during the acquisition, which might cause blurring and biases in the reconstructed maps. In this study, we propose to reconstruct accurate quantitative T1 and T2 maps with reduced blurring compared to the conventional QALAS method using our proposed zero-shot deep subspace reconstruction method (i.e., Zero-DeepSub).Introduction
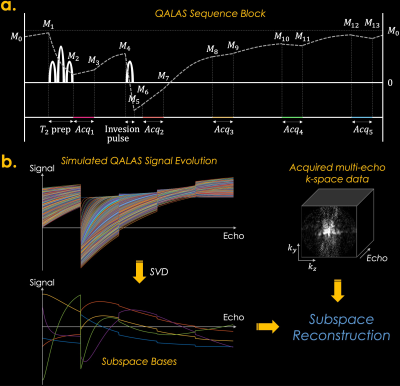
Low-rank subspace/shuffling methods have been powerful for reconstructing time-resolved MRI data and quantitative MRI (qMRI) since they incorporate subspace bases that are calculated from Bloch equations1-3. The 3D-quantification using an interleaved Look-Locker acquisition sequence with T2 preparation pulse (3D-QALAS) has been developed and used for acquiring high-resolution T1, T2, and PD maps from five measurements within each repetition time (TR)4-6. However, when fitting the quantitative maps using a Bloch-simulated dictionary, it assumes that each k-space data is acquired instantly at the first echo of the lengthy echo train, thus neglecting T1 and T2 relaxation during the acquisition, which might cause blurring and biases in the reconstructed T1 and T2 maps. Thus, in this study, we propose to reconstruct QALAS time-series data using a low-rank subspace method and enable more accurate T1 and T2 mapping with reduced blurring compared to the conventional QALAS method. The overall scheme is presented in Fig. 1. Furthermore, we propose a novel zero-shot deep-learning subspace method (i.e., Zero-DeepSub), which combines a scan-specific deep-learning-based reconstruction method7-9 with a low-rank subspace, to further improve the fidelity of multiparametric quantitative MRI.Example Code: https://anonymous.4open.science/r/Zero-DeepSub/
Methods & Experiments
Problem Formulation:Subspace reconstruction can be formulated as follows:
$$\begin{aligned}\underset{\mathbf{x}}{\operatorname{min}}\left\|\mathbf{y}-\mathbf{Ax}\right\|^2_2+\lambda\cal{R}\mathrm{(}\mathbf{x}\mathrm{)}\,,&&(1)\end{aligned}$$
with
$$\begin{aligned}\mathbf{A}=\mathbf{MFC{\Phi}}:\mathbb{C}^{N\times{K}}\to\mathbb{C}^{N\times{C}\times{T}}\,,&&(2)\end{aligned}$$
where $$$\mathbf{y}$$$ denotes the acquired multi-echo and multi-coil k-space data, $$$\mathbf{x}$$$ the denotes the desired subspace coefficient images, and $$$\mathbf{A}$$$ denotes the forward operator that has a k-space sampling matrix $$$\mathbf{M}$$$, Fourier transform $$$\mathbf{F}$$$, coil sensitivity map $$$\mathbf{C}$$$, and subspace bases $$$\mathbf{\Phi}$$$, which transforms the subspace coefficients ($$$\mathbb{C}^{N\times{K}}$$$) into multi-echo and multi-coil k-space data ($$$\mathbb{C}^{N\times{C}\times{T}}$$$). $$$N$$$, $$$K$$$, $$$C$$$, and $$$T$$$ denote the matrix size of the image, number of bases, coils, and echoes. $$$\cal{R\mathrm{(}\cdot\mathrm{)}}$$$ denotes the regularization term and $$$\lambda$$$ denotes the regularization parameter. The l1-wavelet-based regularization is generally used for a subspace reconstruction.
Proposed Model: We propose to use a zero-shot self-supervised learning scheme8 for subspace reconstruction where Eq. 1 with the deep-learning-based regularization10 can be represented as follows:
$$\begin{aligned}\underset{\mathbf{x}}{\operatorname{min}}\left\|\mathbf{y}-\mathbf{MFC{\Phi}x}\right\|^2_2+\lambda\left\|\mathbf{x}-\mathbf{\mathcal{D\mathrm{(}\mathbf{x};\boldsymbol{\theta}\mathrm{)}}}\right\|^2_2\,,&&(3)\end{aligned}$$
where $$$\mathcal{D}$$$ is the convolutional neural network (CNN)-based denoiser with trainable parameters $$$\boldsymbol{\theta}$$$, which can be optimized by minimizing the training loss $$$\mathcal{L_{train}}$$$:
$$\begin{aligned}\underset{\mathbf{\boldsymbol{\theta}}}{\operatorname{min}}\sum_{p=1}^{P}\mathcal{L_{train}}\Big({\mathbf{y}}_{\mathrm{\Lambda}_{p}},\:{\mathbf{A}}_{\mathrm{\Lambda}_{p}}\cal{H}\mathrm{(}{\mathbf{y}}_{\mathrm{\Theta}_{p}},{\mathbf{A}}_{\mathrm{\Theta}_{p}};{\boldsymbol{\theta}}\mathrm{)}\Big)\,,&&(4)\end{aligned}$$
and optimal parameters $$$\boldsymbol{\theta}$$$ can be determined by observing the validation loss $$$\mathcal{L_{val}}$$$:
$$\begin{aligned}\mathcal{L_{val}}\Big({\mathbf{y}}_{\mathrm{\Gamma}},\:{\mathbf{A}}_{\mathrm{\Gamma}}\cal{H}\mathrm{(}{\mathbf{y}}_{\mathrm{\Omega\backslash\Gamma}},{\mathbf{A}}_{\mathrm{\Omega\backslash\Gamma}};{\boldsymbol{\theta}_p}\mathrm{)}\Big)\,,&&(5)\end{aligned}$$
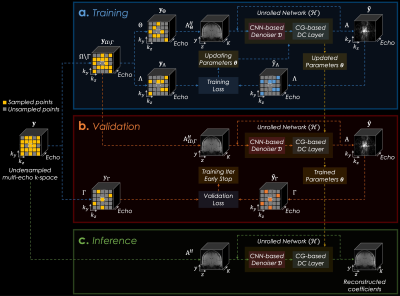
where $$$\cal{H}\mathrm{(}{\mathbf{y}}_{\mathrm{(\cdot)}},{\mathbf{A}}_{\mathrm{(\cdot)}};{\boldsymbol{\theta}_{\mathrm{(\cdot)}}}\mathrm{)}$$$ is the function of the unrolled network using the k-space data $$$\mathbf{y}_{\mathrm{(\cdot)}}$$$, forward model $$$\mathbf{A}_{\mathrm{(\cdot)}}$$$, and trainable parameters $$$\boldsymbol{\theta}_{\mathrm{(\cdot)}}$$$. Here, a k-space sampling strategy is used, which splits the original k-space sampling mask into three different subsets without overlap (i.e., $$$\Omega=\Theta\sqcup\Lambda\sqcup\Gamma$$$) for model training $$$\Theta$$$, training loss $$$\Lambda$$$, and validation loss $$$\Gamma$$$ in each epoch $$$p$$$ ($$$p=1,...,P$$$). The detailed architecture of our proposed model is presented in Fig. 2.
Implementation Details: The CNN-based denoiser consists of 5 layers where each layer has a 3x3 convolutional layer with 128 channels and a leaky ReLU activation with 0.05 negative slope coefficient. The trainable parameter $$$\lambda$$$ was initialized as 0.05. We used conjugate gradients for a data consistency layer with 10 iterations. A total of 4 iterations were performed in the unrolled network. A loss function $$$\cal{L}$$$ was defined as the summation of l1 and l2 norm of the difference between the reconstructed and acquired k-space, which were multiplied with the calculated sampling mask. We made different subsets of sampling masks in each epoch without pre-determined ones using $$$\left|\Omega\right|/\left|\Gamma\right|$$$=0.2 and $$$\left|\Lambda\right|/\left|{\Omega\backslash\Gamma}\right|$$$=0.4. The model was trained with Adam and implemented using Tensorflow.
Acquisition and Simulation: We acquired data from a volunteer using 3D-QALAS sequence on a 3T Prisma scanner with a 32ch head receive array. The parameters are as follows: FOV=240x240x202mm3, matrix size=206x206x176, BW=330Hz/pixel, echo-spacing=5.76ms, turbo factor=128, TR=4.5s, TE=2.29ms, acceleration R=2, and scan time=8m 24s. We retrospectively conducted undersampling with R=2x5 for further validation. For the simulation, we also generated the k-space data using QALAS Bloch-simulation with the same scan parameters.
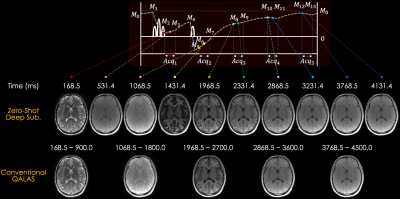
Experiments: We evaluated our proposed Zero-DeepSub by comparing it with 1) conventional QALAS that fits the T1 and T2 maps using original five measurements, 2) subspace reconstruction without regularization, and 3) subspace reconstruction with l1-wavelet regularization. The dictionary was generated with the following T1, and T2 ranges: T1=[300–5000ms] and T2=[10–500ms]. We used 4 bases that could generate the simulated signals within 1.25% errors. The sequence diagram of QALAS is presented in Fig. 1b. We used BART toolbox for estimating coil sensitivity maps and comparison subspace methods11.
Results
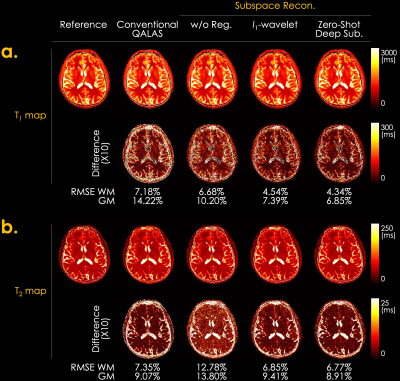
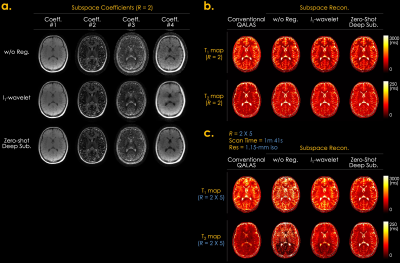
Fig. 3 shows the simulation results of T1 and T2 maps reconstructed using the conventional QALAS and subspace methods without regularization, l1-wavelet regularization, and proposed Zero-DeepSub. The proposed method shows reduced T1 and T2 biases compared to other methods as quantified by root mean squared error (RMSE) of white and gray matter. Fig. 4 presents in vivo subspace coefficients and T1 and T2 maps reconstructed using three different subspace methods. The proposed method shows noise-reduced and sharper coefficients, especially for the third and fourth ones, which result in better T1 and T2 maps. Furthermore, we can resolve interim images within each acquisition with virtual inversion times (TIs) while conventional QALAS only has five reconstructed volumes (Fig. 5).Discussion & Conclusion
In this study, we demonstrated that accurate T1 and T2 maps with reduced blurring can be obtained using the proposed Zero-DeepSub, which combines scan-specific deep-learning reconstruction with low-rank subspace, from 3D-QALAS measurements.Acknowledgements
This work was supported by research grants NIH R01 EB032378, R01 EB028797, R03 EB031175, U01 EB025162, P41 EB030006, U01 EB026996, and the NVidia Corporation for computing support.References
[1] Tamir, J.I., Uecker, M., Chen, W., Lai, P., Alley, M.T., Vasanawala, S.S. and Lustig, M., 2017. T2 shuffling: sharp, multicontrast, volumetric fast spin‐echo imaging. Magnetic resonance in medicine, 77(1), pp.180-195.
[2] Wang, F., Dong, Z., Reese, T.G., Rosen, B., Wald, L.L. and Setsompop, K., 2022. 3D Echo Planar Time-resolved Imaging (3D-EPTI) for ultrafast multi-parametric quantitative MRI. NeuroImage, 250, p.118963.
[3] Cao, X., Liao, C., Iyer, S.S., Wang, Z., Zhou, Z., Dai, E., Liberman, G., Dong, Z., Gong, T., He, H. and Zhong, J., 2022. Optimized multi‐axis spiral projection MR fingerprinting with subspace reconstruction for rapid whole‐brain high‐isotropic‐resolution quantitative imaging. Magnetic Resonance in Medicine, 88(1), pp.133-150.
[4] Kvernby, S., Warntjes, M.J.B., Haraldsson, H., Carlhäll, C.J., Engvall, J. and Ebbers, T., 2014. Simultaneous three-dimensional myocardial T1 and T2 mapping in one breath hold with 3D-QALAS. Journal of Cardiovascular Magnetic Resonance, 16(1), pp.1-14.
[5] Fujita, S., Hagiwara, A., Hori, M., Warntjes, M., Kamagata, K., Fukunaga, I., Andica, C., Maekawa, T., Irie, R., Takemura, M.Y. and Kumamaru, K.K., 2019. Three-dimensional high-resolution simultaneous quantitative mapping of the whole brain with 3D-QALAS: an accuracy and repeatability study. Magnetic resonance imaging, 63, pp.235-243.
[6] Fujita, S., Hagiwara, A., Hori, M., Warntjes, M., Kamagata, K., Fukunaga, I., Goto, M., Takuya, H., Takasu, K., Andica, C. and Maekawa, T., 2019. 3D quantitative synthetic MRI‐derived cortical thickness and subcortical brain volumes: Scan–rescan repeatability and comparison with conventional T1‐weighted images. Journal of Magnetic Resonance Imaging, 50(6), pp.1834-1842.
[7] Yaman, B., Hosseini, S.A.H., Moeller, S., Ellermann, J., Uğurbil, K. and Akçakaya, M., 2020. Self‐supervised learning of physics‐guided reconstruction neural networks without fully sampled reference data. Magnetic resonance in medicine, 84(6), pp.3172-3191.
[8] Yaman, B., Hosseini, S.A.H. and Akcakaya, M., 2021, September. Zero-Shot Self-Supervised Learning for MRI Reconstruction. In International Conference on Learning Representations.
[9] Zhang, M., Xu, J., Arefeen, Y. and Adalsteinsson, E., 2022. Zero-Shot Self-Supervised Learning for 2D T2-shuffling MRI Reconstruction. ISMRM 2022, pp.4051.
[10] Aggarwal, H.K., Mani, M.P. and Jacob, M., 2018. MoDL: Model-based deep learning architecture for inverse problems. IEEE transactions on medical imaging, 38(2), pp.394-405.
[11] Uecker, M., Ong, F., Tamir, J.I., Bahri, D., Virtue, P., Cheng, J.Y., Zhang, T. and Lustig, M., 2015, May. Berkeley advanced reconstruction toolbox. In Proc. Intl. Soc. Mag. Reson. Med (Vol. 23, No. 2486).
Figures