1048
In silico brain data in ADEPT, A Database for MR-Electrical Properties Tomography applications1Department of Radiotherapy, UMC Utrecht, Utrecht, Netherlands, 2Computational Imaging Group for MR Therapy and Diagnostics, UMC Utrecht, Utrecht, Netherlands
Synopsis
Keywords: Electromagnetic Tissue Properties, Electromagnetic Tissue Properties
This work presents ADEPT, A Database for MR-Electrical Properties Tomography applications. ADEPT is an open database that contains simulated MRI field data from electromagnetic simulations on brain models with and without tumor inclusion for EPT reconstructions. These data can serve as common ground to benchmark EPT reconstruction methods developed in different centers and will alleviate the computational burden for the creation of simulated data for training data-driven EPT methods. In the future, the database will be openly accessible and will be extended with phantom and in-vivo data. Other centers are also encouraged to share their data.Introduction
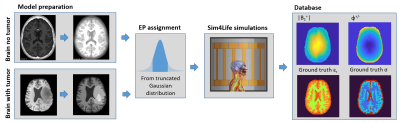
Research developments in Electrical Properties Tomography (EPT) have produced numerous promising reconstruction methods for electrical properties (EP, conductivity σ, and permittivity εr) mapping1. However, direct comparison of reconstruction methods is currently difficult because of a lack of common test data. Furthermore, with the increasing use of data driven reconstruction methods, there is an increasing need for training data, which is computationally expensive to create. To address these issues, we present ADEPT, A Database for EPT reconstructions consisting of simulated data on brain models with and without tumors. Here, we present the setup of the database, the adopted brain models with augmentations, and all the simulated data that have been generated.Methods
At this moment, ADEPT is focused on the brain and consequently includes both healthy brain models (84) and pathological brain models (36) with realistic tumor inclusions.Model creation:
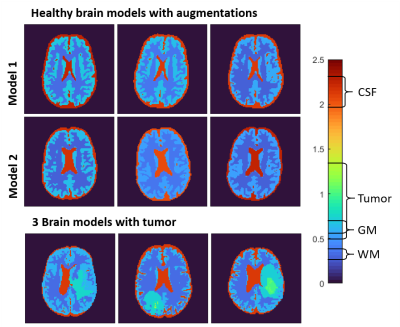
Healthy brain models were generated from pre-segmented, anatomical brain data from BrainWeb2. For the generation of the brain models for ADEPT, different tissues from these pre-segmented models were combined with slicer 3D into 5 tissue classes: White Matter (WM), Gray Matter (GM), Cerebrospinal Fluid (CSF), Muscle and Skull3. Each of these models was augmented 12 times, first employing 3 different geometric augmentations (e.g. model rotations and translations) and second, for each of them, 4 different combinations of tissue EPs (see below).
Brain models with tumor inclusions were created from the BraTS dataset4, which contains T1-weighted images and pre-segmented tumor regions (edema, active tumor, and non-active tumor). For these models, WM, GM and CSF were segmented from the provided T1-weighted images using SPM12 (Functional Imaging Laboratory, University College London, UK). The segmentations were then combined and smoothed into a single volumetric surface model, with tumor segmentation overriding brain tissue. For these models, no geometrical augmentations were performed. Every model was simulated twice, in which different conductivity values were assigned to the tumor area while EPs in WM, GM, and CSF were kept the same.
Assignment of GT EPs:
For the tissue of healthy brain models, conductivity and permittivity were assigned with a random value drawn from a truncated Gaussian distributions with mean EPs values set according to literature: 0.34, 0.59, and 2.14 S/m for conductivity and 53, 73, and 84 for permittivity for respectively WM, GM, and CSF. For the brain models with tumor inclusions, the above reported conductivity and permittivity literature values were used for WM, GM and CSF, while the tumor conductivity values were drawn from a truncated Gaussian distribution with mean values 0.71, 0.90, and 1.20 S/m for edema, non-active tumor, and active tumor, respectively.
EM Simulations:
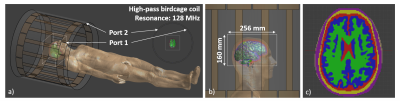
All simulations were performed in Sim4Life (ZMT AG, Zurich, Switzerland) using a 3 T birdcage body coil with quadrature and anti-quadrature mode, in transmission and reception respectively. This allows computations of the transceive phase. Simulation results were exported in a 3D region around the brain of 128x128x80 voxels on a 2 mm isotropic grid. To achieve realistic coil loading the Duke body model was used, where the Duke brain was substituted with the generated brain models5 (Fig 1)
An overview of the entire ADEPT pipeline for in silico data is shown in Fig 2.
Results
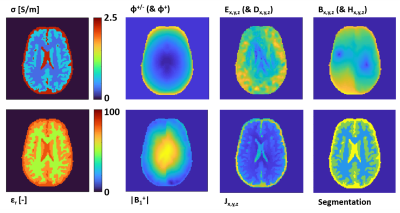
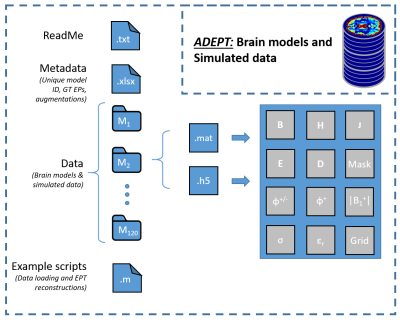
Using the created pipeline, in total 120 brain models are simulated (12 x 7 healthy models and 2 x 18 models with tumor inclusions). Examples of the created models are shown in Fig 3.For each model, the |B1|, φ+, φ+/-, Bx,y,z, Ex,y,z, Jx,y,z, Dx,y,z, Hx,y,z fields are computed and exported. Additionally, the ground truth conductivity and permittivity maps, as well as tissue masks and grid locations are provided. For a single example model an overview of all data saved into ADEPT, is shown in Fig 4.
The structure of the ADEPT database is further shown in Fig 5.
Discussion
To our knowledge, ADEPT is the first open access simulation database for MR-EPT. This has several advantages: first of all, the availability of a common simulated dataset can enable EPT reconstructions on the same data. This makes it possible to compare and benchmark different reconstruction methods. Secondly, it alleviates the computational burden of creating exhaustive databases for training data-driven reconstruction methods. Finally, the availability of data will lower the threshold for research groups to start on research in MR-EPT reconstruction.Yet, ADEPT is still an initial implementation at this point. With additional data (e.g. from other anatomies, using different coils, and different field strengths), the database will become more exhaustive. We therefore encourage other centers to share data for this database as well. Additionally, we plan to further extend this database with the addition of phantom and in-vivo measurements.
In the end, the database will be openly shared and further details will be provided at ISMRM and in the EMTP hub website.
Conclusion
We present ADEPT, a database with in silico brain models and simulated electromagnetic data for MR-EPT reconstructions. This data can be used for comparisons of reconstruction methods and facilitate the development of data driven methods necessitating large amount of data in training.Acknowledgements
This work has been financed by the Netherlands Organisation for Scientific Research (NWO), VENI grant number:18078.References
1. Leijsen et. al. "Electrical properties tomography: A methodological review." Diagnostics. 11.2 (2021): 176. DOI: 10.3390/diagnostics11020176
2. Cocosco, C. A. et. al. "Brainweb: Online interface to a 3D MRI simulated brain database." NeuroImage. 5(4) (1997): S425
3. Fedorov, Andriy, et al. "3D Slicer as an image computing platform for the Quantitative Imaging Network." Magnetic resonance imaging. 30.9 (2012): 1323-1341. DOI: 10.1016/j.mri.2012.05.001
4. B. H. Menze et al. “The multimodal brain tumor image segmentation benchmark (BRATS)." IEEE Transactions on Medical Imaging 34(10) (2015): 1993-2024. DOI: 10.1109/TMI.2015.2502596
5. Christ, A. et al. "The Virtual Family—development of surface-based anatomical models of two adults and two children for dosimetric simulations." Physics in Medicine & Biology. 55, 23–38 (2010).
Figures