1023
MR SIGNATURE MATCHING (MRSIGMA) FOR VOLUMETRIC MRI WITH LESS THAN 300MS LATENCY ON A 1.5T MR-LINAC SYSTEM
Syed Saad Siddiq1, Victor Murray1, Neelam Tyagi1, Pim T.S. Borman2, Bas W. Raaymakers2, Can Wu1, and Ricardo Otazo1,3
1Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Radiotherapy, University Medical Center Utrecht, Utrecht, Netherlands, 3Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States
1Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Radiotherapy, University Medical Center Utrecht, Utrecht, Netherlands, 3Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States
Synopsis
Keywords: Hybrid & Novel Systems Technology, Radiotherapy
The MR-Linac system offers a platform to adapt and monitor radiotherapy of tumors affected by continuous motion in real-time. However, real-time MRI technology is restricted to 2D imaging, which limits performance. MR Signature Matching (MRSIGMA) is a recently developed real-time 3D MRI technique with the MR-Linac as a target. However, MRSIGMA was tested only in a non-real-time scenario. This work develops a real-time implementation of MRSIGMA on the Elekta Unity 1.5T MR-Linac, including raw data transmission to an external computer, deep learning 4D reconstruction and correlation-based matching to demonstrate total imaging latency lower than 300ms.INTRODUCTION
The MR-Linac system offers a platform to adapt and monitor radiation treatment of tumors affected by continuous motion in real-time 1, 2. However, real-time MRI technology is restricted to 2D imaging, which is not able to capture volumetric deformations due to respiratory motion in the chest and abdomen. MR Signature Matching (MRSIGMA) is a recently developed real-time 3D MRI technique that can obtain 3D images with very low imaging latency 3, 4. However, MRSIGMA was only tested as a proof-of-concept on a non-real-time scenario 5. This work develops a real-time implementation of MRSIGMA on the Elekta Unity 1.5T MR-Linac to perform 4D MRI.METHODS
MRSIGMA has two steps: motion learning and signature matching. Motion learning computes a dictionary of 3D motion images and their corresponding signatures using golden-angle radial k-space acquisition and motion-resolved 4D reconstruction. Signature matching acquires signature-only data and finds the closest signature in the motion dictionary. The real-time implementation of MRSIGMA (Figure 1) includes raw data transmission from the 1.5T Unity MR-Linac via 10GBit Ethernet to a high-performance computer (128 cores, 1TB RAM, 2 NVIDIA Ampere 100 GPUs) running motion learning and signature matching pipelines implemented in Python.Data acquisition: T1-weighted golden-angle stack-of-stars acquisition was performed with golden-angle rotation on a healthy volunteer (male, 39 years). For motion learning, 905 angles were continuously acquired with a scan time of 5:35min. The acquisition time of each angle (real-time point) during real-time signature matching was about 250ms.
Raw data transmission: ReconSocket 6 was employed to send each radial k-space line independently to the reconstruction computer. With data acquisition and data transmission working in parallel, latency was minimized (Figure 2).
Motion learning: This step is based on motion-resolved 4D imaging using XD-GRASP 7, which consists of motion detection, motion sorting and 4D reconstruction. The iterative compressed sensing algorithm used in the original implementation of MRSIGMA was replaced by a deep learning reconstruction network named MRI-movienet to minimize computation time. MRI-movienet uses a modified U-net architecture with residual learning blocks to exploit space-time-coil correlations in the image domain without enforcing k-space data consistency. Motion detection computes a respiratory motion signal directly from the central k-space points along kz using principal component analysis, which is classified into 10 amplitude bins. Motion signatures are given by the motion range of the amplitude bins. Raw k-space data are then sorted into 10 motion states. FFT along kz and kx-ky Non-Uniform FFT (NUFFT) transform raw data into aliased 5D images (x-y-z-motion-coil), which are used as input to the MRI-Movienet network to obtain unaliased 4D images.
Signature matching: The distance-based matching algorithm used in the original implementation of MRSIGMA was replaced by a motion projection correlation analysis to reduce computation time. After all kz lines for one angle are received, FFT along kz is performed to obtain a new motion projection or signature. The 3D motion state in the motion dictionary with the signature that presents the highest correlation with respect to the newly acquired motion projection is selected as output.
RESULTS
Figure 2 illustrates a timeline of events with respective latencies. Motion learning was performed in about 341 seconds, of which 335 seconds were acquisition+transmission and only about 5 seconds for 4D reconstruction. Motion detection and motion sorting were completed in 12ms and 483ms respectively. 4D reconstruction by the MRI-Movienet network was completed in only 5 seconds, a significant reduction compared to iterative compressed sensing in the range of tens of minutes. Signature matching latency consists of signature data acquisition of each angle for 250ms and simultaneous signature data transmission to reconstruction computer which only added 3ms ± 2ms. Total signature matching latency was 275ms approximately. Figure 3 displays images from the 4D training dictionary of the volunteer corresponding to their respective motion signature range using the real-time implementation of MRSIGMA. Figure 4 shows the real-time 3D images from signature matching acquired for 60 signatures (18s) with a latency of 300ms.DISCUSSION
Simultaneous data acquisition and transmission minimized the overhead of sending raw data to an external computer. The use of deep learning significantly reduced the time for motion-resolved 4D reconstruction, which in addition to generating the 4D dictionary in less than 5 seconds, can be used to implement 4D MRI in general for other applications in the MR-Linac.CONCLUSION
The combination of parallel data acquisition and data transmission, deep learning-based 4D reconstruction and motion projection correlation matching enabled real-time implementation of the MRSIGMA technique on a Elekta Unity MR-Linac to perform 3D MRI in the abdomen with a total latency lower than 300ms. This development would enable the potential of the MR-Linac system to adapt and monitor treatment of mobile tumors in real-time.Acknowledgements
The work was supported by NIH Grant R01CA255661.References
- Raaymakers, B.W., et al., Integrating a 1.5 T MRI scanner with a 6 MV accelerator: proof of concept. Phys Med Biol, 2009. 54(12): p. N229-37.
- Mutic, S. and J.F. Dempsey, The ViewRay system: magnetic resonance-guided and controlled radiotherapy. Semin Radiat Oncol, 2014. 24(3): p. 196-9.
- Feng, L., N. Tyagi, and R. Otazo, MRSIGMA: Magnetic Resonance SIGnature MAtching for real-time volumetric imaging. Magn Reson Med, 2020. 84(3): p. 1280-1292.
- Kim, N., et al., MR SIGnature MAtching (MRSIGMA) with retrospective self-evaluation for real-time volumetric motion imaging. Phys Med Biol, 2021. 66(21): p. 215009.
- Wu, C., et al., Real-time 3D MRI for Low-Latency Volumetric Motion Tracking on a 1.5 T MR-Linac System. Proc Intl Soc Mag Reson Med 2022; 30:0230.
- Borman, P.T.S., B.W. Raaymakers, and M. Glitzner, ReconSocket: a low-latency raw data streaming interface for real-time MRI-guided radiotherapy. Phys Med Biol, 2019. 64(18): p. 185008.
- Feng, L., et al., XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med, 2016. 75(2): p. 775-88.
Figures
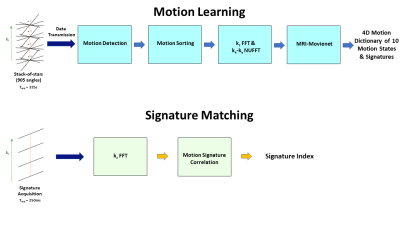
Figure
1:
Motion
Learning data
processing pipeline (highlighted in blue) generates 4D motion dictionary
consisting of 3D images of 10 motion states. Signature
Matching data
processing pipeline (highlighted in yellow) selects one of 10 motion states in
the training dictionary based on new signatures acquired from MR-Linac
in real-time.
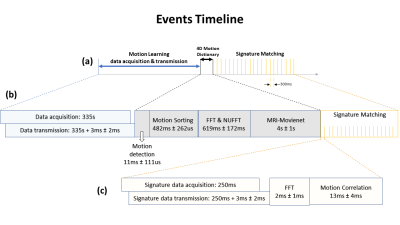
Figure
2:
a) Timeline
of events of the MRSIGMA technique, b)
Motion learning data acquisition, transmission and
processing latencies are shown , c) Signature
matching step
latencies are shown.
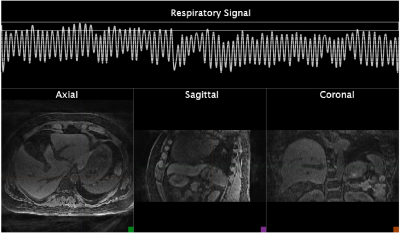
Figure 3 (GIF video): Motion images from 4D dictionary with corresponding
respiratory motion signature range depicted at the top. Varying displacements
of organs moving in and out of the axial, sagittal and coronal views can be
observed.
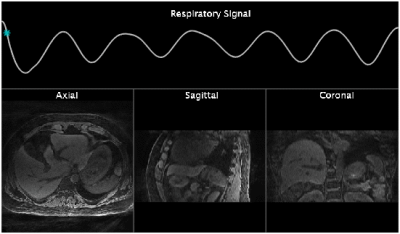
Figure 4 (GIF video): Real-time 4D imaging of a healthy volunteer using
MRSIGMA for 60 consecutive time points.
DOI: https://doi.org/10.58530/2023/1023