0990
MRI reconstruction via Data-Driven Markov Chains with Joint Uncertainty Estimation: Extended Analysis1University Medical Center Göttingen, Göttingen, Germany, 2Institute of Biomedical Imaging, Graz University of Technology, Graz, Austria, 3German Centre for Cardiovascular Research (DZHK), Partner Site Göttingen, Göttingen, Germany, 4Cluster of Excellence "Multiscale Bioimaging: from Molecular Machines to Networks of Excitable Cells'' (MBExC), University of Göttingen, Göttingen, Germany
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction
The application of generative models in MRI reconstruction is shifting researchers’ attention from the unrolled reconstruction networks to the probabilistically tractable iterations and permits an unsupervised fashion for medical image reconstruction.Introduction
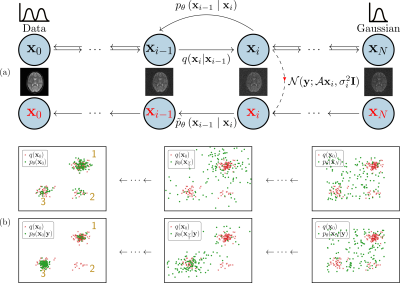
The application of generative models in MRI reconstruction is shifting researchers' attention from the unrolled reconstruction networks to the probablistically tractable iterations and permits an unsupervised fashion for medical image reconstruction [1-4].We formulate the image reconstruction problem from the perspective of Bayesian inference, which enables efficient sampling from the learnt posterior probability distributions [1-2]. Different from conventional deep learning-based MRI reconstruction techniques, samples are drawn from the posterior distribution given the measured k-space using the Markov chain Monte Carlo (MCMC) method. The data-driven Markov chains are the reverse of a diffusion process that transfers the distribution of data to a known distribution (See Figure 1). Since the generative model learned from a image database is independent of the forward operator, this means the same pretrained models can be applied to k-space acquired with different sampling schemes or receive coils. Here, we present additional results in terms of the uncertainty of reconstruction, the transferability of learned information, and the comparison in the fastMRI challenge.
Theory
In order to compute the posterior probability $$$p(\mathbf{x}|\mathbf{y})$$$ for the image $$$\mathbf{x}$$$ given the measured k-space data $$$\mathbf{y}$$$, we need to modify the learned reverse process. We can achieve this by multiplying each of the intermediate distributions $$$p(\mathbf{x}_i)$$$ with the likelihood term $$$p(\mathbf{y}|\mathbf{x}_i)$$$ (see Figure 1). We use $$$\tilde{p}\left(\mathbf{x}_{i}\right)$$$ to denote the resulting sequence ofintermediate distributions\begin{equation} \tilde{p}\left(\mathbf{x}_{i}\right)\propto p\left(\mathbf{x}_{i}\right) p(\mathbf{y}|\mathbf{x}_i)\end{equation}up to the unknown normalization constant. Following [5], the transition from $$$\mathbf{x}_{i+1}$$$ to $$$\mathbf{x}_i$$$ of the modified reverse process is\begin{equation} \tilde{p}\left(\mathbf{x}_{i} \mid \mathbf{x}_{i+1}\right)\propto p\left(\mathbf{x}_{i} \mid \mathbf{x}_{i+1}\right) p(\mathbf{y}|\mathbf{x}_i). \end{equation} For the measument model of k-space, we express it as \begin{equation} p(\mathbf{y}|\mathbf{x}_i)=\mathcal{CN}(\mathbf{y};\mathcal{A}\mathbf{x}_i, \sigma_\eta^2\mathbf{I}).\end{equation}The sampling at each intermediate distribution of Markov transitions is performed with the unadjusted Langevin Monte Carlo [6] \begin{equation} \mathbf{x}_i^{k+1} \leftarrow \mathbf{x}_i^{k} + \frac{\gamma}{2}\nabla_{\mathbf{x}_i}\log \tilde{p}(\mathbf{x}_{i}^{k}\mid\mathbf{x}_{i+1}) + \sqrt{\gamma}\mathbf{z}, \label{eq:ld}\end{equation}where $$$\mathbf{z}$$$ is standard complex Gaussian noise $$$\mathcal{CN}(0, \mathbf{I})$$$. With the learned reverse process $$$p_{\boldsymbol{\theta}}\left(\mathbf{x}_i \mid \mathbf{x}_{i+1}\right)$$$, the log-derivative of $$$\tilde{p}_{\boldsymbol{\theta}}(\mathbf{x}_{i}\mid\mathbf{x}_{i+1})$$$ with respect to $$$\mathbf{x}_i$$$ is\begin{equation} \nabla_{\mathbf{x}_i}\log \tilde{p}_{\boldsymbol{\theta}}(\mathbf{x}_{i}\mid\mathbf{x}_{i+1}) = \nabla_{\mathbf{x}_i}\log p_{\boldsymbol{\theta}}\left(\mathbf{x}_i \mid \mathbf{x}_{i+1}\right) + \nabla_{\mathbf{x}_i}\log p(\mathbf{y}|\mathbf{x}_i),\end{equation}where\begin{equation} \nabla_{\mathbf{x}_i}\log p_{\boldsymbol{\theta}}\left(\mathbf{x}_i \mid \mathbf{x}_{i+1}\right) = \frac{1}{\tau_{i+1}^2}\left(\sigma_{i+1}^{2}-\sigma_{i}^{2}\right) \mathbf{s}_{\boldsymbol{\theta}}\left(\mathbf{x}_{i+1}, i\right)\end{equation}and \begin{align} \nabla_{\mathbf{x}_i}\log p(\mathbf{y}|\mathbf{x}_i) & = -\frac{1}{\sigma_\eta^2}(\mathcal{A}^{H} \mathcal{A}\mathbf{x}_i-\mathcal{A}^{H}\mathbf{y}_0).\end{align} For more details about how to learn reverse process and create measurement operator $$$\mathcal{A}$$$, please check our preprint [2].Methods
In our preprint [2], we proposed Algorithm 1 for simulating samples from the posterior $$$p(\mathbf{x}|\mathbf{y})$$$ in which the score network [7] is used to construct Markov transition kernels. The following experiments are selected for this abstract. Codes and data to reproduce all experiments are made available in [8].1. Multi-coil reconstruction
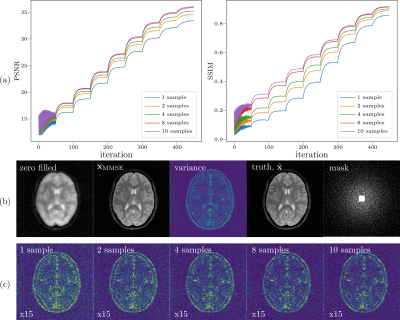
Multi-channel data points from k-space are randomly picked with variable density poisson-disc and the central 20$$$\times$$$20 region is fully acquired. The acquisition mask covers 11.8% k-space and the corresponding zero-filled reconstruction is shown Figure 2.We initialized 10 chains and the $$$\mathbf{x}_\mathrm{MMSE}$$$ was computed using different number of samples. The parameters in Algorithm 1 are $$$\mathrm{K}=50, \mathrm{N}=10, \lambda=13$$$.To visualize the process of sampling, we use peak-signal-noise-ratio (PSNR in dB) and similarity index (SSIM) as metrics to track intermediate samples.The comparisons are made between the magnitude of $$$\mathbf{x}_\mathrm{MMSE}$$$ and the ground truth $$$\tilde{\mathbf{x}}$$$ after normalized with $$$\ell_2$$$-norm.
2. Transferability
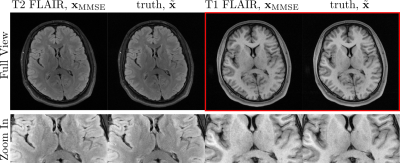
To investigate the transferability of learned prior information from T2 FLAIR images to other contrasts, we acquired T1-weighted (TR=2000ms, TI=900ms, TE=9ms) and T2-weighted (TR=9000ms, TI=2500ms, TE=81ms) FLAIR k-space data using a 2D multi-slice turbo spin-echo sequence with a 16-channel head coil at 3T (Siemens, 3T Skyra). The parameters in Algorithm 1 are $$$\mathrm{K}=2, \mathrm{N}=70, \lambda=20$$$.
3. Comparison to fastMRI challenge
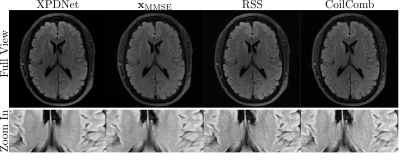
As a comparison to the unrolled neural network, the XPDNet is selected as the reference which ranked 2nd in the fastMRI challenge. Two networks were trained for acceleration factors 4 and 8, using retrospectively under-sampleddata from the fastMRI dataset [9] with equidistant Cartesian masks [10] and the trained models are available on [11]. For the proposed method, the parameters in Algorithm 1 are $$$K=4, N=90, \lambda=20$$$.
Results and Discussions
1. Multi-coil reconstructionComparing with the ground truth, the variance map mainly reflects the edge information, which can be interpreted by the uncertainty that is introduced by the undersampling pattern used in k-space where many high frequency data points are missing but the low frequency data points are fully acquired. In contrast to the single coil unfolding, the local spatial information from coil sensitivities reduces the uncertainties of missing k-space data. Moreover, error maps qualitatively correspond to the variance map, with larger errors in higher variance regions as shown in Figure 2.
2. Transferability
Figure 3 shows a score network trained with T2 FLAIR contrast used to reconstruct a T1 FLAIR image in comparison to a T2 FLAIR image. No loss can be observed.
3. Comparison to fastMRI challenge
As discussed in [12], the ground truth matters when computing metrics for the reconstruction. The RSS and Coil-combined are used as ground truth for XPDnet and $$$\mathbf{x}_\mathrm{MMSE}$$$.
Conclusion
The proposed reconstruction method combines concepts from machine learning, Bayesian inference and image reconstruction. In the setting of Bayesian inference, the image reconstruction is realized by drawing samples from the posterior term $$$p(\mathbf{x}|\mathbf{y})$$$ using data driven Markov chains, providing a minimum mean square reconstruction and uncertainty estimation. Furthermore, this method shows good performance in terms of transferabilities of contrasts and sampling patterns.Acknowledgements
No acknowledgement found.References
[1] Luo, G., Heide, M., & Uecker, M. Using data-driven Markov chains for MRI reconstruction with Joint Uncertainty Estimation. ISMRM Annual Meeting 2022, In Proc. Intl. Soc. Mag. Reson. Med. 30;0298 (2022)
[2] Luo, G., Heide, M., & Uecker, M. (2022). MRI Reconstruction via Data Driven Markov Chain with Joint Uncertainty Estimation. arXiv preprint arXiv:2202.01479.
[3] Jalal A., Arvinte M., Daras G., Price E., Dimakis Alexandros G., Tamir J. (2021). Robust Compressed Sensing MRI withDeep Generative Priors. In: Ranzato M., Beygelzimer A., DauphinY., Liang P.S., Vaughan J. Wortman, eds. Advances in Neural Infor-mation Processing Systems, :14938–14954 Curran Associates, Inc..
[4] Chung H., Ye Jong C. (2022, May) Score-based diffusion models foraccelerated MRI. Medical Image Analysis. 2022;80:102479.
[5] Sohl-Dickstein, J., Weiss, E., Maheswaranathan, N., & Ganguli, S. (2015, June). Deep unsupervised learning using nonequilibrium thermodynamics. In International Conference on Machine Learning (pp. 2256-2265). PMLR.
[6] Douc, R., Moulines, E., Priouret, P., & Soulier, P. (2018). Markov chains (p. 757). Cham, Switzerland: Springer International Publishing.Chicago
[7] Song, Y., & Ermon, S. (2019). Generative modeling by estimating gradients of the data distribution. Advances in Neural Information Processing Systems, 32.
[8] https://github.com/mrirecon/spreco
[9] Zbontar, J., Knoll, F., Sriram, A., Murrell, T., Huang, Z., Muckley, M. J., ... & Lui, Y. W. (2018). fastMRI: An open dataset and benchmarks for accelerated MRI. arXiv preprint arXiv:1811.08839.
[10] Facebookresearch. (n.d.). Facebookresearch/fastmri: A large-scale dataset of both raw MRI measurements and clinical MRI images. Retrieved November 10, 2022, from https://github.com/facebookresearch/fastMRI/
[11] Zaccharieramzi. (n.d.). Zaccharieramzi/fastmri-reproducible-benchmark: Try several methods for MRI reconstruction on the FASTMRI dataset. Retrieved November 10, 2022, from https://github.com/zaccharieramzi/fastmri-reproducible-benchmark
[12] Arvinte, M., Tamir, J. The truth matters: A brief discussion on MVUE vs. RSS in MRI reconstruction. ESMRMB 2021.
Figures