0988
Constructing age-specific MRI brain templates based on a uniform healthy population across life span with transformer1Columbia University, New York, NY, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Brain
Image registration is currently employed to assist with diagnostic tasks such as neurodegenerative disease diagnosis. Aging also affects the brain, and our knowledge of age-related brain diseases is currently encumbered by age-induced bias; understanding the mechanisms of neurodegenerative diseases requires a deeper understanding of aging. Thus we have devised a pipeline for generating age-specific templates using a novel deep-learning algorithm detailed in a separate report. Our results show qualitative changes in brain morphology across age groups, and tissue segmentation was performed on each template to calculate volumetric changes in brain matter across time.Introduction
Age-related volumetric changes in brain tissue are well-documented and are especially profound during adulthood1. In spite of these discoveries, studies aimed at identifying volumetric brain changes tend to use scans of young adults’ brain tissue for segmentation reference which biases the results1. The segmentation studies typically classify brain matter into three categories: gray matter (GM), white matter (WM), and cerebral spinal fluid (CSF) and utilize automated processes to guide the segmentation via a priori reference data2,3. These a priori insights are derived from anatomical templates commonly available for scientific use. Naturally, the quality of the a priori insights is a function of the anatomical biases inherent to the analysis. Studies prove that specific templates reduce anatomic biases in MRI analysis4. It has been asserted that this bias reduction is caused by the more precise mappings afforded to study-specific templates1. Reports have also indicated that high-resolution nonlinear spatial normalization tools can further reduce anatomical bias5. Unfortunately, we found no reports that use deep learning to automate the nonlinear approximations required to create age-specific templates via image registration. We propose a novel method for generating age-specific templates of human MRI scans using a network we’ve detailed in another report. The iterative strategy for producing the templates has been proven effective in previous works. The accuracy of our deep learning model, coupled with its novel use of transformers and inverse consistency to maintain topology, has created a powerful tool for visualizing and quantifying changes in brain morphology and volume across time.Methods
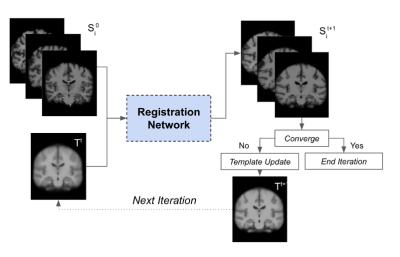
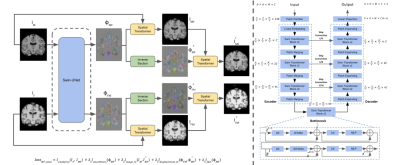
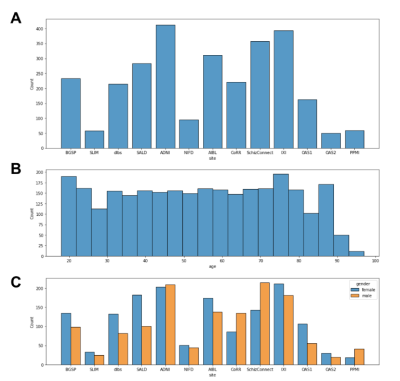
Fig. 1 is an overview of the age-specific template construction pipeline inspired by Sanchez et al. First an initial template (T0) is generated by averaging the subject scans, which have already been registered to the MNI152 template before averaging. Then we adopt a learnable deformable registration model to register all the subject scans to the initial template, as shown in Fig. 2, and thus generate a new set of registered subject scans. The registration’s performance is measured by the coefficient of determination (R2) between the initial template and each registered subject scan. We only use subject scans with the top 5% R2 values and take the R2-weighted average over the scans; this decreases convergence time and alleviates bias. This new template is then used as the reference in the next iteration. The iteration continues until the R2 value between the new template and the previous template has reached a threshold (eg. R2 = 0.999). Upon iteration convergence, we finally register all the subject scans to this template and generate the final template by taking the R2-weighted average over all the registered subject scans. The final result is the desired age-specific template.The neuroimaging data12 were collected from 13 different sites with a total of 2852 T1-weighted brain MRI scans are used in this study. As illustrated in Fig. 3A, our dataset exhibits an approximately uniform distribution over both lifespan (18 - 97 years old) and gender (male/female). We further divide these subject scans into 10 age groups (18-25, 26-33, 34-41, 42-49, 50-57, 58-65, 66-73, 74-81, 82-89, 90-97) according to age. In our data preprocessing pipeline, we first corrected the bias field inhomogeneity with the N4 bias field correction procedure and affine-registered the raw whole-head scans to the MNI152 unbiased template8,9. Subsequently, the Brain Extraction Tool10 was applied to achieve skull-stripping. After that, we affine-registered these scans to the MNI152 unbiased template.
Results
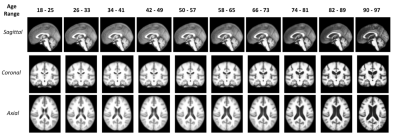
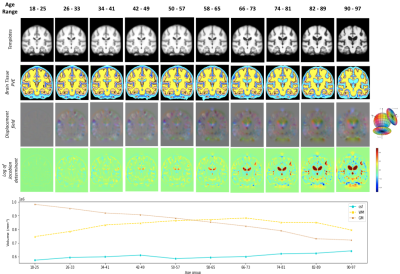
Fig. 4 shows ten age-specific templates constructed using our pipeline. Fig. 5 shows both the qualitative and quantitative results to indicate how the overall brain structure changes over the lifespan. The coronal slice of the brain templates has been used for visualization in Fig. 5.Discussion
With our deep learning-based age-specific template construction pipeline, we are successful in generating a set of templates that can meaningfully capture the morphological changes over the human lifespan. Although the templates shown in Fig. 4 are the preliminary templates generated by our pipeline, clear morphological changes (especially in the ventricle of the brain) are observable. Fig. 5 indicates that while gray matter volume decreases monotonically, white matter volume actually increases until middle age (~45 years), at which point white matter decreases. This result is comparable to the previous outcomes11. As we can see in the white matter volume plot, the volume of white matter in the age group 90-97 is higher than the one in the age group 18-25. This is likely induced by the shortage of subject scans available in the 90-97 age group which increases the bias.Conclusion
Our work provides a new deep learning-based age-specific template construction pipeline that can meaningfully represent the neuroanatomical changes occurring over lifespan. The cycle inverse-consistent transformer-based deformable registration model achieves high registration accuracy due to the use of Swin-UNet. The inverse consistency and topology preservation properties of the transformation are guaranteed by a comprehensive consideration of the inverse-consistent loss. In the future, we will further explore the structural changes in subjects by implementing tissue segmentation on the registered subject scans.Acknowledgements
No acknowledgement found.References
1. Fillmore PT, Phillips-Meek MC, Richards JE. Age-specific MRI brain and head templates for healthy adults from 20 through 89 years of age. Front Aging Neurosci. 2015 Apr 8;7:44. doi: 10.3389/fnagi.2015.00044. PMID: 25904864; PMCID: PMC4389545.
2. Zhang, Y. Y., Brady, M., and Smith, S. (2001). Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Trans. Med. Imaging 20, 45–57. doi: 10.1109/42.906424
3. Zijdenbos, A. P., Forghani, R., and Evans, A. C. (2002). Automatic ‘‘pipeline’’ analysis of 3-D MRI data for clinical trials: application to multiple sclerosis. IEEE Trans. Med. Imaging 21, 1280–1291. doi: 10.11
4. Good, C. D., Johnsrude, I. S., Ashburner, J., Henson, R. N. A., Friston, K. J., andFrackowiak, R. S. J. (2001). A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage 14, 21–36. doi: 10.1006/nimg.2001.078609/tmi.2002.806283
5. Callaert, D. V., Ribbens, A., Maes, F., Swinnen, S. P., and Wenderoth, N. (2014). Assessing age-related gray matter decline with voxel-based morphometry depends significantly on segmentation and normalization procedures. Front. Aging Neurosci. 6:124. doi: 10.3389/fnagi.2014.00124
6. Junyu Chen, Eric C. Frey, Yufan He, William P. Segars, Ye Li, Yong Du, TransMorph: Transformer for unsupervised medical image registration, Medical Image Analysis, Volume 82, 2022, 102615, ISSN 1361-8415, https://doi.org/10.1016/j.media.2022.102615.
7. Dale, A. M., Fischl, B., and Sereno, M. I. (1999). Cortical surface-based analysis.I. Segmentation and surface reconstruction. Neuroimage 9, 179–194. doi: 10.1006/nimg.1998.0395
8. Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001 Jun;5(2):143-56. doi: 10.1016/s1361-8415(01)00036-6. PMID: 11516708.
9. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002 Oct;17(2):825-41. doi: 10.1016/s1053-8119(02)91132-8. PMID: 12377157.
10. Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002 Nov;17(3):143-55. doi: 10.1002/hbm.10062. PMID: 12391568; PMCID: PMC6871816.
11. Sowell, E., Peterson, B., Thompson, P. et al. Mapping cortical change across the human life span. Nat Neurosci 6, 309–315 (2003). https://doi.org/10.1038/nn1008
12. Feng, X., Lipton, Z. C., Yang, J., Small, S. A., Provenzano, F. A., Alzheimer’s Disease Neuroimaging Initiative, Australian Imaging Biomarkers and Lifestyle flagship study of ageing, & Frontotemporal Lobar Degeneration Neuroimaging Initiative (2020). Estimating brain age based on a uniform healthy population with deep learning and structural magnetic resonance imaging. Neurobiology of aging, 91, 15–25. https://doi.org/10.1016/j.neurobiolaging.2020.02.009
Figures