0987
Refining synthetic training data improves image quality transfer for ultra-low-field structural brain MRI1Computer Science, University College London, London, United Kingdom, 2Gates Foundation, Seattle, WA, United States, 3University of Stavanger, Stavanger, Norway, 4University of Ibadan, Ibadan, Nigeria, 5University College London, London, United Kingdom, 6Great Ormand Street Hospital for Children, London, United Kingdom
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Low-Field MRI, super-resolution, Hyperfine
Computer vision methods can be used for image quality transfer (IQT) to address the poor contrast, decreased resolution and increased noise observed in MR images acquired at ultra-low magnetic fields. Current methods have been shown to produce high-quality synthetic outputs for low-field (~0.5T) but not ultra-low-field (~0.05T) field images. Moreover, these methods do not adapt well to the presence of abnormal morphology (e.g. lesions). Here we introduce a new approach to ultra-low field IQT that improves on previous methods and is adaptive to the presence of synthetic lesions.
INTRODUCTION
Image quality transfer (IQT) is a computer vision-based approach to generate high-quality estimates of low-quality images. The usual methodology is to synthesise low-field training data by degrading high-field-acquired image data and to use this paired data to train either machine learning1 or deep learning2,3 models to generate a synthetic high-quality output.SynthSR2,4 is a software that generates a high-resolution T1-weighted image equivalent to any input scan. While SynthSR has demonstrated excellent results on synthetic low-field (<1T), data2, it performs less well on image data acquired at ultra-low (<0.05T) fields (Fig 1). Moreover, SynthSR does not adapt well to abnormal morphology such as the presence of lesions (Fig 2). This may be related to the fact that SynthSR is designed to be contrast agnostic. This is achieved by manipulating separately the grey-scale voxel intensities of segmented brain structures to generate a low-field synthetic image, which is mapped to an authentic high-field T1-weighted image. The result is that the model learns to devalue information related to grey-scale values, and instead over-relies on features related to brain morphology. Because of this, brain lesion images may be problematic as these are generally characterised as a change in the expected grey-scale value in a region rather than a change in the large-scale morphology of the brain. As a result, SynthSR-enhanced images lesions appear to be "filled in" to reflect the general expected morphology in that region.
To avoid these problems, we generate synthetic low-resolution training images by degrading the high-resolution image as a whole (i.e. not treating structures separately). We also make use of K-space sub-sampling, gamma transforms and noise addition when degrading high-resolution images to generate synthetic low-resolution training images that qualitatively approximate ultra-low-field images. Finally, we adopt a ResUNet5 architecture (rather than a standard UNet2,3) to learn the mapping between synthetic low-resolution and high-resolution images. When applied to authentic ultra-low-field acquired MR brain images, our results demonstrate an increase in image resolution, decrease in noise and an improvement in tissue contrast. We further demonstrate that our approach is adaptable to synthetic lesions.
METHODS
DataBias-field corrected T1-weighted images from the Human Connectome Project6 (HCP) were used as training data (n=100). Images were of dimensions [260x260x311], with voxels of side 0.7mm3. Test data were acquired on a Hyperfine system (0.064T). The T1-weighted images had dimensions [120x146x36], with voxels of side 1.6x1.6x5mm3. To evaluate model adaptability to abnormal morphology, we additionally generated a synthetic lesion dataset by manually drawing "lesions" (areas of consistent voxel intensity) on sample images from the HCP dataset. These were subsequently degraded to generate a synthetic low-resolution lesion image.
IQT Pipeline
In order to perform IQT we first degraded high-resolution image data to generate synthetic low-quality data. These paired data were then patched to smaller volumes and filtered to remove mean-zero patches before being used to train a ResUNet model.
Image Preprocessing
The preprocessing pipeline is outlined in Fig 3. Synthetic low-resolution data was generated by first skull-stripping image data, standardising and subsequently cubing image dimensions to [256x256x311]. Thereafter image resolution was reduced by a factor [2x2x8] to reflect reduced dimensions of ultra-low field images. The images were FFT transformed into K-space and sub-sampled. Noise was added before transformation back to image space and applying Gaussian kernel for blurring. The images were then restored to original dimensions using cubic spline interpolation, and a mask applied to remove noise from non-brain voxels. The hyperparameters controlling the degree of K-space subsampling, noise and blurring were generated probabilistically to provide a diverse range of training images of varying quality.
For testing, the resolution of Hyperfine images was increased using cubic spline interpolation. The images were then patched and the trained model applied to each patch. All patches were concatenated to generate a final IQT image.
Training
Training images were patched into volumetric slices of side [256x256x3]. Patches were filtered to exclude those with mean zero. A 4-layer ResUNet architecture4 was implemented in PyTorch and run on Google Colab GPU. Due to computational limitations, the number of filters in each layer was set at [32, 64, 120, 180]. Early stopping criteria was used to end training. The Adam optimizer was used to minimize the L1 loss which has been shown to produce visually sharper boundaries and facilitate training by escaping local minima.7
RESULTS
Results of the trained model applied to authentic ultra-low-field image data are reported in Fig. 4. They show improved tissue contrast, decreased noise and increased resolution. However, the use of thin volumetric patches leads to image artefacts and decreased tissue detail in out-of-plane image slices. This may be addressed in future studies by adopting cubic image patches for training.Qualitatively, the trained model performed well when applied to synthetic low-resolution lesion data (Fig 5), with the resulting IQT’d volume retaining the synthetic lesion.
DISCUSSION & CONCLUSION
Image synthesis for training can play a key role in determining the type of features learned by models designed for IQT. Improvements in synthetic low-resolution image generation and model architecture can significantly benefit the performance of IQT on images acquired at ultra-low field strengths. The results presented here should be validated on external datasets.Acknowledgements
No acknowledgement found.References
1 Alexander DC, Zikic A, Ghosh A, Tanno R, Wottschel V, Zhang J, Kaden E, Dyrby TB, Sotiropoulos SN, Zhang H, Criminisi A. Image quality transfer and applications in diffusion MRI. NeuroImage (2017)
2 Iglesias JE, Billot B, Balbastre Y, Tabari A, Conklin J, RG Gonzalez, Alexander DC, Golland P, Edlow B, Fischl B. Joint super-resolution and synthesis of 1 mm isotropic MP-RAGE volumes from clinical MRI exams with scans of different orientation, resolution and contrast. NeuroImage (2021)
3 Lin H, Figini M, Tanno R, Blumberg SB, Kaden E, Ogbole G, Brown BJ, D’Arco F, Carmicharl DW, Lagunju I, Cross HJ, Fernandez-Reyes D, Alexander DC. Deep learning for low-field to high-field MR: Image quality transfer with probabilistic decimation simulator. MICCAI (2019)
4 Iglesias JE, Schleicher R, Laguna S, Billot B, Schaefer P, McKaig B, Goldstein JN, Sheth KN, Rosen MS, Kimberly WT. Accurate super-resolution low-field brain MRI. arxiv.org/abs/2202.03564
5 Jha D, Smedsrud PH, Riegler MA, Johansen D, de Lange T, Halvorsen P, Johansen HD. ResUNet++: An advanced architecture for medical image segmentation. IEEE ISM (2019)
6 Glasser MF, Sotiropoulos SN, Wilson A, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M, WU-Minn HCP Consortium. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage (2013).
7 Ooi YK, Ibrahim H. Deep learning algorithms for single image super-resolution: A systematic review. Electronics (2021).
Figures
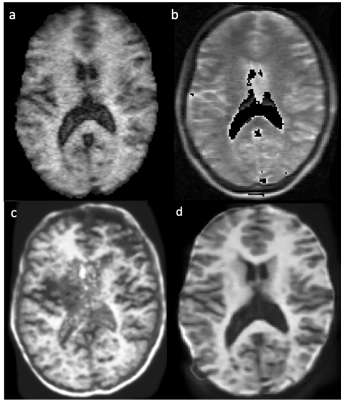
Figure 1: (a, b) Original Hyperfine-acquired T1-weighted and T1-weighted image data respectively. (c) Results of SynthSR applied to (a) only. SynthSR is not optimised for single image input at ultra-low-field strengths. Due to over-reliance on learned morphological features, a skull artefact is introduced in the generated output. (d) SynthSR is optimised for ultra-low-field data using a combined input of T1 and T2 images. SynthSR output based on combined (a) and (b).
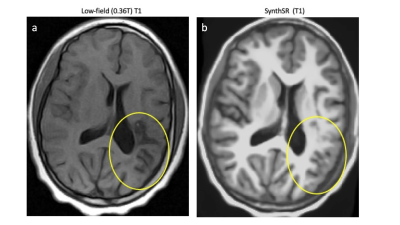
Figure 2: (a) White matter lesion on T1-weighted low-field MRI. (b) Results of SynthSR applied to (a). The lesion appears to be “filled in”. This may be a result of how SynthSR training data is generated, leading to an over-reliance on morphological features at the expense of grey-scale information.
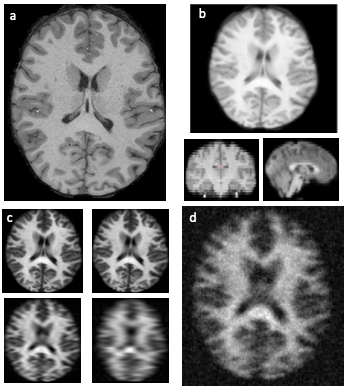
Figure 3: (a) Original HCP image. (b) Dimensions of image are reduced using cubic spline interpolation. (c) Fast-Fourier Transform applied to transform image to K-space for sub-sampling. (d) Noise is added, and image is blurred. Thereafter image is restored to original dimensions and a brain mask is applied to remove background noise.
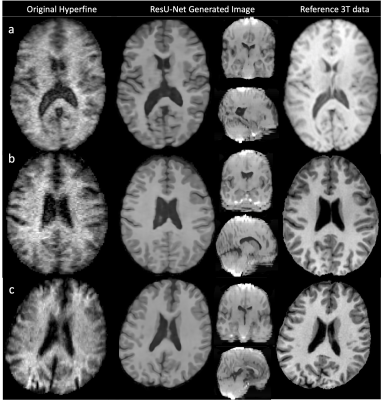
Figure 4: Three examples of Hyperfine-acquired T1-weighted brain images (first column). Axial slices through the ResU-Net generated output (second column) shows improved tissue contrast, reduced noise and increased image resolution, and compares well to the reference 3T image (third column). However, the corresponding coronal and sagittal planes (second column) show image artefacts and poor morphological detail. Switching to cubic patching during training may address this.
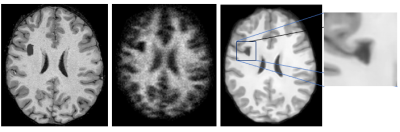
Figure 5: (a) Original HCP sample image with synthetic lesion. (b) Corresponding synthetic low-resolution image based on (a). (c) Results of trained model applied to (b). The “lesion” is maintained and not filled-in.