0986
A novel deep multimodal contrastive network for early prediction of cognitive deficits using multimodal MRI and clinical data1Imaging Research Center, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 2Department of Computer Science, University of Cincinnati, Cincinnati, OH, United States, 3The Perinatal Institute, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Brain Connectivity, Contrastive Learning, Cognitive Deficits
Deep learning has shown promising results in early predicting cognitive deficits in very preterm infants using multimodal brain MRI data, acquired soon after birth. Prior methods focus on the feature fusion of different modalities but ignore the latent high-order feature similarity information. In this study, we propose a novel deep multimodal contrastive network using T2-weighted structural MRI (sMRI), diffusion tensor imaging (DTI), resting-state functional MRI (rsfMRI), and clinical data to predict later cognitive deficits. Our proposed model significantly improved predictive power compared to other peer models for early diagnosis of cognitive deficits.Summary of Main Findings
Compared to other state-of-the-art contrastive models, our novel deep multimodal contrastive network model was able to achieve the best prediction performance with a balanced accuracy of 83.4% and an AUC of 0.82.INTRODUCTION
The prevalence of cognitive deficits remains high for very preterm infants. A precise prognostic model is desirable to address the challenge of early prediction of cognitive impairments in very preterm infants. Recent studies show that the integration of multimodal MRI data with deep learning techniques is more effective than using a single-modality MRI for the early prediction of cognitive deficits1-4. However, prior methods usually focus on the feature fusion of different modalities but ignore the high-order information among modalities and subjects. In this study, we proposed a novel deep multimodal contrastive network for early prediction of cognitive deficits using multimodal brain MRI acquired at term-corrected age in very preterm infants by utilizing the latent higher-order feature similarities among patient-wise modality and class-wise modality.METHODS
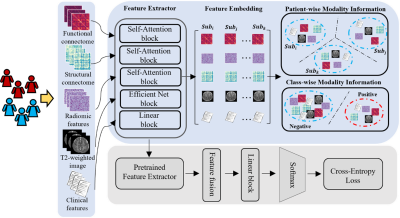
We developed our model using a regional prospective cohort of very preterm infants from the Cincinnati Infant Neurodevelopment Early Prediction Study (CINEPS)5. All infants with known congenital brain anomalies or severe perinatal injury were excluded, resulting in 395 subjects from the CINPES cohort. All subjects were imaged during unsedated sleep on the same 3T Philips Ingenia scanner and 32-channel receiver head coil at 39-44 weeks postmenstrual age. Each subject was assessed at 2 years corrected age using the Bayley Scales of Infant and Toddler Development, 3rd Ed. (Bayley-III) test. Bayley-III Cognitive scores ranged from 40 to 145 in the cohort. We dichotomized subjects into two groups: the low-risk group (score >85) and the high-risk group (85).An overview of our study is shown in Figure 1. We included three modalities of brain MRI data, including T2-weighted structural MRI (sMRI), diffusion tensor imaging (DTI), resting-state functional MRI (rsfMRI), and perinatal clinical data collected prior to neonatal intensive care unit discharge. The original T2-weighted MRI 2D images were processed using the developing Human Connectome Project (dHCP) pipeline6-8 to segment whole brain images into 87 regions of interest (ROIs), from which we extracted a total of 100 agnostic radiomic features using PyRadiomics pipeline9. We then preprocessed DTI and rsfMRI data using the corresponding dHCP pipelines and further constructed brain structural connectome and functional connectome from these modalities, respectively. Finally, we organized data as predictive features for our model. After data preprocessing, we obtained 5 different features/inputs.
Our proposed model was designed to equip 5 feature extractors to take 5 feature types from each subject (Figure 1). Specifically, we applied self-attention mechanisms to learn brain functional connectome, structural connectome, and radiomic features. We applied a pre-trained EfficientNet10 to extract anatomical features from T2-weighted images. We used a fully connected network for clinical features. Next, we design two pretext contrastive learning tasks to extract feature embeddings from 5 feature modalities. The first pretext task is to learn the patient-wise modality information by clustering the feature types of an individual patient, and the second task is to learn the class-wise modality information by clustering the patient with the same class label. These two pretext tasks largely increase the training samples for the deep learning models, mitigating the inadequate data issue for model training in medical applications. Finally, we fine-tuned the pre-trained network to solve the downstream task (i.e., risk stratification of cognitive deficits) in a supervised manner. We evaluated our proposed model and compared it with other peer contrastive methods using 10-fold cross-validation.
RESULTS
Our model was able to achieve a balanced accuracy of 83.4%, a sensitivity of 82.5%, a specificity of 84.3%, and an AUC of 0.82, outperforming other state-of-the-art contrastive learning models (Table 1). In addition, our model also achieved better prediction performance than the single-modality model (Table 2).DISCUSSION AND CONCLUSION
In this study, we proposed a novel deep multimodal contrastive network to predict later cognitive deficits in very preterm infants using multimodal MRI data at term-corrected age. The results showed that our proposed model outperformed other peer models, demonstrating the effectiveness of the predictive power of our designed approach. In future studies, we will externally validate the proposed model using an external cohort.Acknowledgements
This work was supported by the National Institutes of Health [R01-EB029944, R01-EB030582, R01-NS094200, and R01-NS096037]; Academic and Research Committee (ARC) Awards of Cincinnati Children's Hospital Medical Center. The funders played no role in the design, analysis, or presentation of the findings.References
1. He, L., Li, H., Chen, M., Wang, J., Altaye, M., Dillman, J. R., & Parikh, N. A. (2021). Deep multimodal learning from MRI and clinical data for early prediction of neurodevelopmental deficits in very preterm infants. Frontiers in neuroscience, 15.
2. Ball, G., Aljabar, P., Nongena, P., Kennea, N., Gonzalez‐Cinca, N., Falconer, S., ... & Edwards, A. D. (2017). Multimodal image analysis of clinical influences on preterm brain development. Annals of neurology, 82(2), 233-246.
3. Li, X., Jia, M., Islam, M. T., Yu, L., & Xing, L. (2020). Self-supervised feature learning via exploiting multi-modal data for retinal disease diagnosis. IEEE Transactions on Medical Imaging, 39(12), 4023-4033.
4. Stoyanov, D., Taylor, Z., Carneiro, G., Syeda-Mahmood, T., Martel, A., Maier-Hein, L., ... & Madabhushi, A. (Eds.). (2018). Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings (Vol. 11045). Springer.
5. Tamm, L., Patel, M., Peugh, J., Kline-Fath, B. M., Parikh, N. A., Cincinnati Infant Neurodevelopment Early Prediction Study (CINEPS) Group. (2020). Early brain abnormalities in infants born very preterm predict under-reactive temperament. Early Human Development, 144, 104985.
6. Makropoulos, A., et al., The developing human connectome project: A minimal processing pipeline for neonatal cortical surface reconstruction. Neuroimage, 2018. 173: p. 88-112.
7. Tustison, N. J., Avants, B. B., Cook, P. A., Zheng, Y., Egan, A., Yushkevich, P. A., & Gee, J. C. (2010). N4ITK: improved N3 bias correction. IEEE transactions on medical imaging, 29(6), 1310-1320.
8. Makropoulos, A., Gousias, I. S., Ledig, C., Aljabar, P., Serag, A., Hajnal, J. V., ... & Rueckert, D. (2014). Automatic whole brain MRI segmentation of the developing neonatal brain. IEEE transactions on medical imaging, 33(9), 1818-1831.
9. Van Griethuysen, J. J., Fedorov, A., Parmar, C., Hosny, A., Aucoin, N., Narayan, V., ... & Aerts, H. J. (2017). Computational radiomics system to decode the radiographic phenotype. Cancer research, 77(21), e104-e107.
10. Tan, M., & Le, Q. (2019, May). Efficientnet: Rethinking model scaling for convolutional neural networks. In International conference on machine learning (pp. 6105-6114). PMLR.
Figures