0969
Cluster analysis of VERDICT MRI for identification of tumor subregions with distinct histological features1Department of Medical Radiation Sciences, Institute of Clinical Sciences, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden, 2Department of Medical Physics and Biomedical Engineering, Sahlgrenska University Hospital, Gothenburg, Sweden
Synopsis
Keywords: Diffusion/other diffusion imaging techniques, Cancer, VERDICT, preclinical, histology
Characterization of tumor tissue may aid in grading and assessment of cancer treatment. Time-dependent diffusion MRI can provide non-invasive estimates of parameters relating to tissue microstructure in vivo. In this study, a cluster analysis was performed to group parameters estimated by the time-dependent diffusion MRI model VERDICT. Parameter cluster maps were compared with maps derived from classification of histological images. The results showed good agreement between VERDICT cluster maps and histology maps, indicating that the method shows potential in identifying tumor subregions of distinct morphological features.Introduction
Characterization of cancer tissue can facilitate the cancer treatment process by providing tumor grading and biomarkers for assessment of treatment effect, thus providing support for planning and reassessment of the treatment strategy.Time-dependent diffusion MRI (dMRI) allows for non-invasive probing of the tissue microstructure in vivo by making the MR signal sensitive to microstructural restrictions of water diffusion. The Vascular, Extracellular, and Restricted Diffusion for Cytometry in Tumors (VERDICT)1 model can be fit to data acquired at different diffusion times and diffusion weightings to provide estimates of cell radius index (R) and volume fraction index of the intracellular space (fIC).
Cluster analysis allows for grouping of the estimated VERDICT parameters by e.g., Gaussian fitting, to find distinguishable clusters of parameter combinations. This method may enable identification of tumor subregions of distinct histological features that are not apparent when observing parameters individually. Differentiation of such regions could facilitate tumor grading and assessment of treatment effect, as well as providing a tool for non-invasive in vivo analysis of tissue features in research.
The aim of this study was to identify tumor subregions using cluster analysis of VERDICT parameter maps, and to compare them with maps derived from histological analysis of irradiated tumor tissue in a mouse model of human neuroendocrine tumor.
Methods and materials
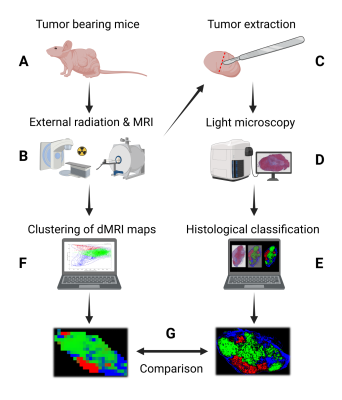
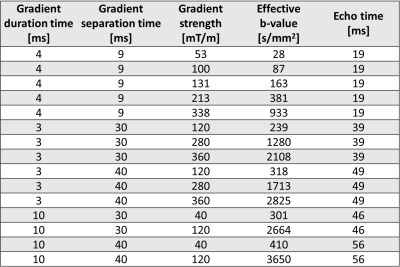
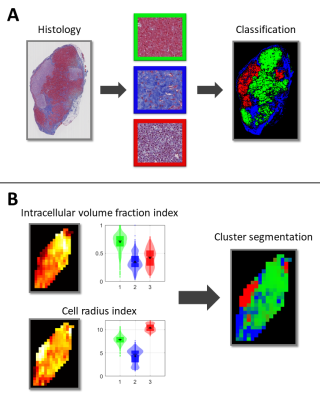
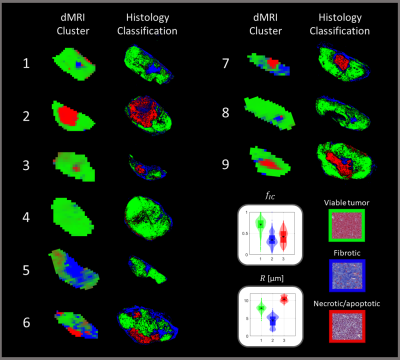
The workflow of the study is outlined in Figure 1. Experiments were conducted on BALB/c nude mice of the human SI-NET model GOT1 (n=9). Tumors were irradiated externally to an absorbed dose of 8 Gy using a 6-MV photon beam. Fifteen days after the treatment the tumors were imaged using a dMRI protocol designed for VERDICT analysis (Table 1). MRI scans were acquired using a 7T MR system (Bruker, Biospec, MRI GmbH, Ettlingen, Germany) with 400×400 μm2 in-plane resolution and 500 μm slice thickness.Following the MRI scans the animals were sacrificed and the tumors were extracted, fixated, and sectioned. Tumor sections through the same plane as the MRI images were stained with Masson’s trichrome (MT) to stain collagen in fibrotic tissue, cell nuclei, and cytoplasm, and imaged using a whole slide light microscopy imager (Leica Biosystems, Germany). A Random Forest pixel classification algorithm was trained on a subset of the MT images to create segmented maps of three distinct tissue types: viable, fibrotic, and necrotic/apoptotic tumor tissue (Figure 2). The algorithm was trained using the Ilastik software2.
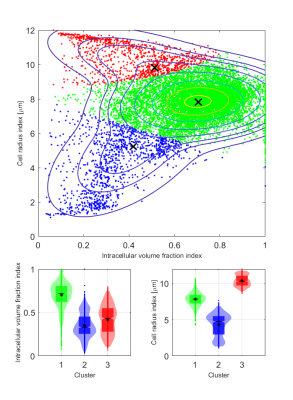
The VERDICT model was fitted to dMRI data using the AMICO framework to estimate R and fIC3. The diffusion coefficient of the intracellular, extracellular extravascular, and vascular space, as well as the velocity dispersion of the blood flow were fixed to 1×10-9m2/s, 1.5×10-9m2/s, 1.75×10-9m2/s, and 0.6x10-3 m/s respectively to make the model fit more robust. A Gaussian mixture model was fitted to the distribution of R and fIC parameter values from all voxels in all tumors using an expectation-maximization algorithm, as has been similarly done before with the IVIM model4. The model had three components to match the number of distinct tissue types defined on the MT images. Soft cluster maps were generated for the central MRI slice of each tumor based on the fit (Figure 3). All model fitting was done using MATLAB (R2020a, MathWorks, Natick, MA).
The study was approved by the Gothenburg Ethical Committee on Animal Research.
Results and discussion
The Gaussian mixture model fit resulted in clusters of distinctively different parameter values (Figure 3). fIC was high for one cluster and low for the other two while R showed substantially different values for all clusters. The classification algorithm generated colormaps which matched well with the histology and was able to accurately distinguish the defined tissue types (Figure 2).Overall, the comparison between the dMRI-based cluster maps and the histology-based classification maps showed good agreement (Figure 4). A perfect spatial match is not expected due to histological tissue deformation during the fixation steps. However, the cluster maps appeared to distinguish the defined tissue types in the histology maps well, indicating the potential use of cluster analysis of VERDICT parameters to obtain information on histological features in tumors.
The cluster with high fIC showed the best spatial agreement with the viable tumor tissue class (Figures 3 & 4), suggesting that more microstructural restriction is present in areas of densely packed cells compared to fibrotic and necrotic/apoptotic tissue. Furthermore, although the clusters that matched best with fibrotic and necrotic/apoptotic tissue showed similar fIC values between one another, their R values differed substantially with fibrotic tissue showing a lower R. This indicates that diffusion-restrictive structures in fibrotic tissue may be smaller compared to those found in necrotic/apoptotic tissue, and that the R parameter contains information on specificity between the two tissue types which is not found in fIC.
Conclusion
Cluster analysis of the VERDICT MRI parameters R and fIC shows potential in identifying tumor tissue subregions of distinct morphological features in the studied tumor model. The method may thus provide a means to non-invasively study oncologically relevant features of the tumor microenvironment in vivo.Acknowledgements
This study was funded by grants from the Swedish Cancer Society, the Swedish Research Council, the King Gustav V Jubilee Clinic Cancer Research Foundation, BioCARE – a National Strategic Research Program at the University of Gothenburg, the Swedish state under the agreement between the Swedish government and the county councils, the ALF agreement, the Sahlgrenska University Hospital Research Funds, the Assar Gabrielsson Cancer Research Foundation, the Adlerbertska Research Foundation, the Herbert & Karin Jacobsson Foundation, the Royal Society of Arts and Sciences in Gothenburg (KVVS), and the Wilhelm and Martina Lundgren Research Foundation.
Figure 1 was created with BioRender.com
References
1. Panagiotaki E, Walker-Samuel S, Siow B, et al. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res 2014;74(7):1902–1912.
2. Berg S, Kutra D, Kroeger T, et al. Ilastik: Interactive Machine Learning for (Bio)Image Analysis. Nat Methods 2019;16(12):1226–1232.
3. Bonet-Carne E, Johnston E, Daducci A, et al. VERDICT-AMICO: Ultrafast fitting algorithm for non-invasive prostate microstructure characterization. NMR Biomed 2018:e4019 doi: 10.1002/nbm.4019.
4. Jalnefjord O, Montelius M, Arvidsson J, et al. Data-driven identification of tumor subregions based on intravoxel incoherent motion reveals association with proliferative activity. Magn Reson Med 2019;82(4):1480–1490.
Figures