0963
A multi-compartment model for pathological connectomes1Diffusion Imaging and Connectivity Estimation (DICE) Lab, Department of Computer Science, University of Verona, Verona, Italy, 2Translational Imaging in Neurology (ThINK), Department of Biomedical Engineering, University of Basel, Basel, Switzerland, 3Sherbrooke Connectivity Imaging Laboratory (SCIL), Département d’Informatique, Université de Sherbrooke, Sherbrooke, QC, Canada, 4Department of Neuroscience, Rehabilitation, Ophthalmology, Genetics, Maternal and Child Health (DINOGMI), University of Genoa, Genoa, Italy, 5Research Center for Clinical Neuroimmunology and Neuroscience Basel (RC2NB), University of Basel, Basel, Switzerland, 6Department of Neurology, MS Center, University Hospital Basel, Basel, Switzerland
Synopsis
Keywords: Diffusion/other diffusion imaging techniques, Brain Connectivity
The white matter is the complex system of neuronal fibers in the brain. Any disruption to this circuitry may lead to a wide range of neurological diseases and, thus, it is fundamental to be able of detecting pathological conditions at early stages. State-of-the-art methods for studying brain connectivity assume constant properties along the fibers, but this assumption is not valid in pathological conditions that locally affect the tissue, e.g. multiple sclerosis. Here, we present a model for “multi-compartment connectomes” to explicitly consider the presence of focal lesions during the estimation of connectivity and show its effectiveness using realistic numerical simulations.Introduction
Our community has long been interested in the study of structural connectivity, a.k.a. connectome, in case of pathology1–9. State-of-the-art methods to perform quantitative tractography analyses assume consistent microstructural properties along fiber pathways10–16, but in the presence of focal lesions, this assumption is violated and introduces a bias in the estimated connectomes4–9. For instance, it was demonstrated that the connectivity of bundles affected by lesions is usually misestimated6,8; however, as illustrated in Figure 1, the situation is more complex than that because, in global estimation approaches, even a single misestimation can trigger a cascade of events, in which also bundles that are not affected by lesions may be overestimated in a sort of compensatory mechanism. Notably, this overall misestimation of connectivity may drastically reduce the sensitivity of connectome-based methods to detect pathological conditions. In this study, we introduce the concept of “multi-compartment connectome”, which is a novel modeling approach to accurately map the structural connectivity also in the presence of focal pathology by explicitly considering the lesions during the estimation process.Methods
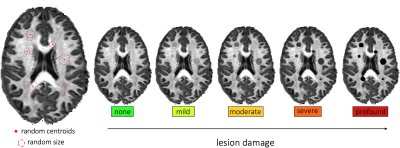
We extended the Convex Optimization Modeling for Microstructure Informed Tractography (COMMIT) framework10 to include an additional tissue compartment in voxels affected by lesions with the aim to explicitly quantify the local tissue damage. Standard COMMIT estimates one coefficient per streamline which represents the cross-sectional area of the fibers. Using the additional information from the lesions compartment, we have the possibility to refine these coefficients and selectively reduce the cross-sectional area of the streamlines passing through lesions, proportionally to the corresponding damage; this procedure was inspired by the adage “A chain is only as strong as its weakest link” recently discussed in 4–9. Finally, connectomes are computed as usual by summing the (possibly reduced) cross-sectional areas of the streamlines belonging to each bundle.We tested our model in 44 HCP healthy subjects (http://www.humanconnectomeproject.org) and, for each, we artificially created a lesion mask by modeling lesions as spheres (Figure 2); centroids were randomly placed within the WM and then randomly dilated until 5% of WM coverage was reached, as typically found e.g. in multiple sclerosis17. To simulate tissue damage, we scaled the MR signal measured in voxels inside lesions according to 5 levels of degeneracy: “none” (i.e., healthy subject); “mild” (25% signal reduction); “moderate” (50%); “severe” (75%), and “profound” (100%). For the sake of simplicity, we used as signal a fiber density scalar map, as done in previous studies, but the model can be generalized to the full dMRI data or other modalities11,12,16.
We compared the percentage change of the two approaches in the connectivity estimation (i.e. “without” and “with” lesion modeling) to assess their sensitivity to detect disruptions in the estimated connectomes. We focused our analysis on three network measures typically used in clinical applications: mean strength, efficiency, and modularity.
Results
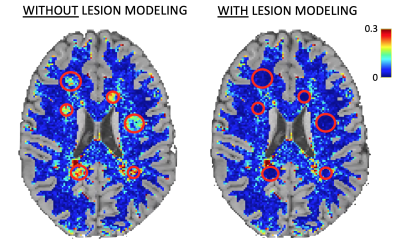
As expected, when lesions are not explicitly modeled, high fitting errors are observed in pathological voxels, whereas the proposed multi-compartment connectome model fits the data accurately also inside the lesions (Figure 3). We can thus be more sensitive to pathological changes even at low levels of degeneracy compared to the standard method (plots are reported in Figure 4, while percentage changes are reported in Figure 5).Discussion
Current state-of-the-art methods for estimating structural connectivity assume constant microstructural properties along the streamlines. Hence, they are not suitable to cope with localized signal alterations due to focal lesions10–16; e.g. it was demonstrated that the connectivity of fibers passing through lesions is typically misestimated. To tackle this problem, an extension of SIFT214 was recently proposed which scales the cross-sectional area of “pathological bundles” according to the signal loss9. However, since lesions are not explicitly modeled during the estimation process, the fit is still biased and results in a compensatory mechanism which leads to an overestimation of the “unaffected bundles” (Figure 1). As a consequence, the sensitivity to neurodegeneration of current methods is limited.Our results show that our multi-compartment connectome model can be successfully applied also to patient connectomes, as it explicitly models focal lesions and does not introduce bias into connectivity estimates, furthermore it is able to detect pathological conditions at early stages of degeneration. Despite promising, our results are based on realistic numerical simulations and only represent a proof-of-concept study which we believe will stimulate research in this direction. For instance, we used tractographies reconstructed from healthy subjects’ data, but it is known that in pathological voxels also the fiber orientations might be affected, and not only the signal magnitude, which may have important consequences on the sensitivity of the estimated connectomes. Future research will be dedicated to testing our model in more complex lesion configurations as well as to evaluating its effectiveness in a real population of patients with focal lesions, e.g. multiple sclerosis.
Conclusion
We introduced the novel concept of “multi-compartment connectome” which, for the first time, allows accurate mapping of the structural brain connectivity also in the presence of focal pathology. This proof-of-concept study opens exciting new possibilities to better understand the underlying mechanisms of neurodegenerative diseases starting from early stages.Acknowledgements
No acknowledgement found.References
1. Griffa A, Baumann PS, Thiran JP, Hagmann P. Structural connectomics in brain diseases. Neuroimage. Published online 2013. doi:10.1016/j.neuroimage.2013.04.056
2. Fornito A, Zalesky A, Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16(3):159-172.
3. Fleischer V, Radetz A, Ciolac D, et al. Graph Theoretical Framework of Brain Networks in Multiple Sclerosis: A Review of Concepts. Neuroscience. Published online 2019. doi:10.1016/j.neuroscience.2017.10.033
4. Sarwar T, Ramamohanarao K, Zalesky A. Mapping connectomes with diffusion MRI: deterministic or probabilistic tractography? Magn Reson Med. 2019;81(2):1368-1384. doi:10.1002/mrm.27471
5. Smith RE, Calamante F, Connelly A. Mapping connectomes with diffusion MRI: Deterministic or probabilistic tractography? Magn Reson Med. 2020;83(3):787-790. doi:10.1002/mrm.27916
6. Zalesky A, Sarwar T, Ramamohanarao K. A cautionary note on the use of SIFT in pathological connectomes. Magn Reson Med. 2020;83(3):791-794. doi:10.1002/mrm.28037
7. Smith RE, Calamante F, Connelly A. Notes on “A cautionary note on the use of SIFT in pathological connectomes.” Magn Reson Med. 2020;84(5):2303-2307. doi:10.1002/mrm.28266
8. Zalesky A, Sarwar T, Kotagiri R. SIFT in pathological connectomes: Follow-up response to Smith and colleagues. Magn Reson Med. 2020;84(5):2308-2311. doi:10.1002/mrm.28412
9. Smith RE, Calamante F, Gajamange S, Kolbe S. Modulation of white matter bundle connectivity in the presence of axonal truncation pathologies. Published online 2020:0-4.
10. Daducci A, Dal Palù A, Lemkaddem A, Thiran J-PP. COMMIT: convex optimization modeling for microstructure informed tractography. IEEE Trans Med Imaging. 2014;34(1):246-257. doi:10.1109/TMI.2014.2352414
11. Schiavi S, Ocampo-Pineda M, Barakovic M, et al. A new method for accurate in vivo mapping of human brain connections using microstructural and anatomical information. Sci Adv. 2020;(a). doi:10.1126/sciadv.aba8245
12. Schiavi S, Lu PJ, Weigel M, et al. Bundle myelin fraction (BMF) mapping of different white matter connections using microstructure informed tractography. Neuroimage. 2022;249(January):118922. doi:10.1016/j.neuroimage.2022.118922
13. Smith RE, Tournier JD, Calamante F, Connelly A. SIFT: Spherical-deconvolution informed filtering of tractograms. Neuroimage. 2013;67:298-312. doi:10.1016/j.neuroimage.2012.11.049
14. Smith RE, Tournier J-D, Calamante F, Connelly A. SIFT2: Enabling dense quantitative assessment of brain white matter connectivity using streamlines tractography. Neuroimage. 2015;119:338-351.
15. Pestilli F, Yeatman J, Rokem A, Kay K, Takemura H, Wandell B. LiFE: Linear Fascicle Evaluation a new technology to study visual connectomes. J Vis. 2014;14(10):1122.
16. Ocampo-Pineda M, Schiavi S, Rheault F, et al. Hierarchical microstructure informed tractography. Brain Connect. 2021;11(2):75-88.
17. Ouellette R, Treaba CA, Granberg T, et al. 7 T imaging reveals a gradient in spinal cord lesion distribution in multiple sclerosis. Brain. 2020;143(10):2973-2987.
Figures
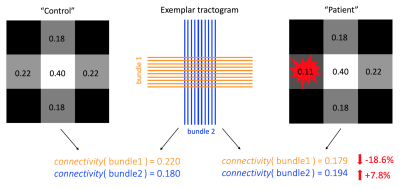
Application of a state-of-the-art structural connectomics method on proof-of-concept data. On the left, we report the values of the bundles’ connectivity and the corresponding fiber-density scalar map in the absence of focal lesions, while on the right we introduce a focal lesion. The effect of the lesion is reflected not only on the bundles that pass through it (underestimation, orange) but, potentially, ALSO on all the others (overestimation, blue).
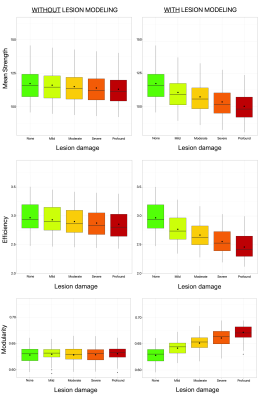
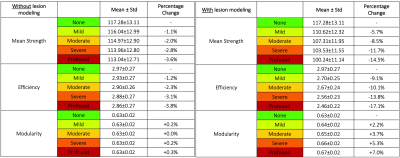
Percentage change of the global network measures between each lesion damage (i.e. mild, moderate, severe, profound) and the corresponding reference values (i.e. none), defined as: ((None – Lesion damage) / None) × 100%.