0950
VarNet-based Simultaneous Multislice Reconstruction for Low Noise Amplification and Flexible Calibration1College of Health Science and Environmental Engineering, Shenzhen Technology University, Shenzhen, China, 2University Research Facility in Behavioral and Systems Neuroscience, The Hong Kong Polytechnic University, Hong Kong, China
Synopsis
Keywords: Image Reconstruction, Parallel Imaging
In this study, we combine the readout-concatenation framework with VarNet based deep learning method to achieve SMS reconstruction of low noise amplification. It can flexibly handle arbitrary slice aliasing patterns with in-plane acceleration. Moreover, the modified network architecture allows coil calibration using a prescan of different contrasts, which is preferred in many scenarios.Introduction
Simultaneous multislice imaging (SMS) has rapidly developed into a major imaging acceleration technique. In addition to traditional SENSE1, GRAPPA2 and Slice-GRAPPA3 (SG) reconstruction methods, deep learning-based methods may also be adapted to SMS image reconstruction. Traditional reconstruction methods may suffer from high noise amplification at high acceleration factors, whereas many deep learning methods are not flexible. For example, vanilla VarNet4 requires coil sensitivity calibration internally from the same scan, while some other methods require ESPIRIT7 calibration in advance, which may be suboptimal and inconvenient.In this study, we combine the readout-concatenation framework with VarNet based deep learning method to achieve SMS reconstruction of low noise amplification. It can flexibly handle arbitrary slice aliasing patterns with in-plane acceleration. Moreover, the modified network architecture allows coil calibration using a prescan of different contrasts, which is preferred in many scenarios.
Methods
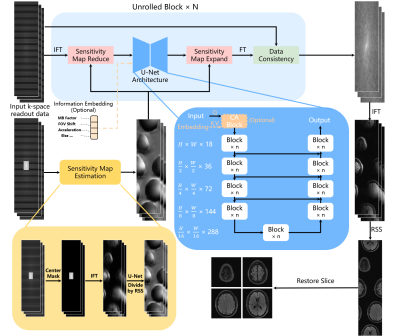
VarNet-based SMS reconstructionThe proposed SMS reconstruction method is based on the VarNet network and readout-concatenation9 framework as shown in Fig. 1. The internal U-structure module can be a simple U-Net (denoted as VarNet) or more advanced NAFNet5 structure (denoted as NAF-VarNet).
Unlike the original VarNet method, we separately feed the SMS data and the autocalibration signal (ACS) data to the network. This enables external calibration using ACS data from a prescan to estimate coil sensitivity, which is desirable for faster clinical examination and in fact the routine practice for fMRI and DWI applications.
Note that prior information of the scan, such as the multiband (MB) factor, FOV shift, and intra-plane acceleration (R) factor, can be optionally used in the cross-attention module (CA block in Figure 1). We denoted this improved model as NAF-VarNet-Plus.
Experiments on fastMRI
We synthesized SMS data from fastMRI8 multicoil brain data. The training and validation sets were as officially divided. We first used the CAIPIRINHA6 method to introduce a FOV shift for each slice that was simultaneously excited, setting the shift to be the inverse of the MB factor. Then the data were summed along slice direction and reorganized to form readout-concatenated data. The calibration region size was set to 30 x 30.
Training details
All VarNet-based methods were trained with mixed MB factors ranging from 2 to 5 randomly in combination with in-plane acceleration of 1, 4, and 8. Due to the GPU memory limit, we set the number of VarNet cascade modules to 3, and reduced the number of NAFNet blocks to 2 per layer for fair comparison. Network weights were iteratively optimized by minimizing the SSIM loss and Charbonnier loss. SSIM is a structural similarity index, while Charbonnier loss is more robust than L1 loss and can better handle outliers.
Experiments on low-field data with external calibration
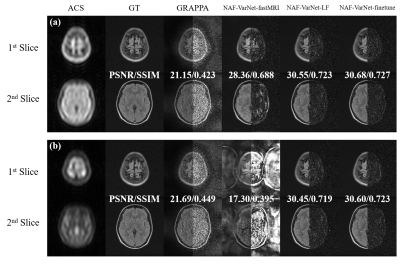
To investigate the feasibility of external coil calibration using prescan, we acquired FLAIR data (matrix size=256×198, FOV=240×240, TR/TE/TI=7500/98/1655, ETL=11) and T2-weighted data (matrix size=256×195, FOV=240×240, TR/TE =5500/122, ETL=13) on a 0.3T MRI scanner (Oper-0.3, Ningbo Xingaoyi) from 150 human subjects. SMS acceleration was introduced retrospectively as in the fastMRI experiments to the FLAIR data and the T2w data were used for coil sensitivity calibration. We compared three VarNet models: one trained solely on fastMRI (denoted as NAF-VarNet-fastMRI), one solely trained on the low-field data (denoted as NAF-VarNet-LF), and one finetuned on the low-field data after fastMRI pretraining (denoted as NAF-VarNet-finetune).
Results
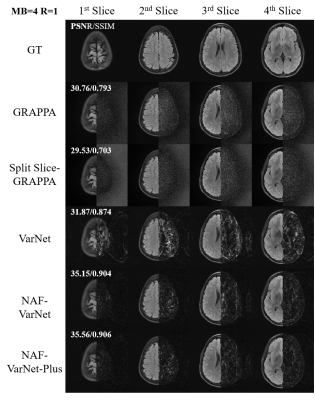
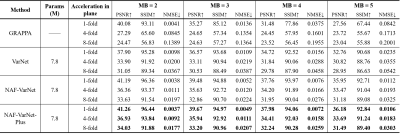
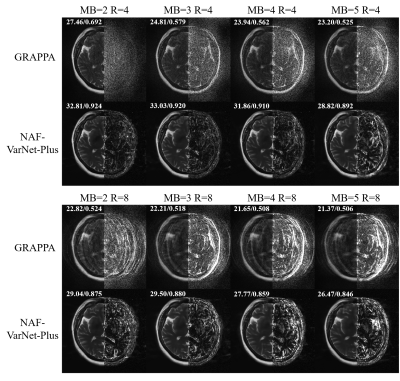
As visually plotted in Figure 2 and quantitatively measured in Figure 3, the proposed VarNet-based methods vastly reduced the noise amplification compared to GRAPPA. Particularly, NAF-VarNet and NAF-VarNet-Plus performed better than the baseline VarNet (simple UNet-based). Figure 4 shows the reconstructed images of NAF-VarNet-Plus and GRAPPA under different multiband (MB) and in-plane acceleration (R) factors, where substantial improvement by NAF-VarNet-Plus can be observed. Figure 5 demonstrated the feasibility of using external ACS data of T2 weighting to reconstruct FLAIR SMS data, for which fine-tuning was found necessary because the fastMRI brain dataset did not include such multicontrast data.Discussion and Conclusion
In this abstract, we present a VarNet-based SMS reconstruction method that is flexible to combine with in-plane acceleration. Experimental results show that this method greatly improves image quality compared with traditional parallel imaging. It not only performed robustly across datasets but also enables external calibration using prescans. Nonetheless, there are some limitations of the current study. First, we have not evaluated the methods on prospectively accelerated data. Second, the large image size of readout concatenated data causes very high GPU memory usage. Last, we did not compare it with alternative approaches using ESPIRiT as the coil calibration method. However, as has been previously studied4, our end-to-end approach is more convenient and potentially has higher limits.Acknowledgements
This study is supported in part by Natural Science Foundation of Top Talent of Shenzhen Technology University (Grants No. 20200208 to Lyu, Mengye) and the National Natural Science Foundation of China (Grant No. 62101348 to Lyu, Mengye)References
1. Pruessmann, Klaas P., et al. "SENSE: sensitivity encoding for fast MRI." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 42.5 (1999): 952-962.
2. Griswold, Mark A., et al. "Generalized autocalibrating partially parallel acquisitions (GRAPPA)." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 47.6 (2002): 1202-1210.
3. Setsompop, Kawin, et al. “Blipped‐controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g‐factor penalty.” Magnetic resonance in medicine 67.5 (2012): 1210-1224.
4. Sriram, Anuroop, et al. "End-to-end variational networks for accelerated MRI reconstruction." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2020.
5. Chen, Liangyu, et al. "Simple baselines for image restoration." arXiv preprint arXiv:2204.04676 (2022). 6. Breuer, Felix A., et al. "Controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA) for multi‐slice imaging." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 53.3 (2005): 684-691.
7. Uecker, Martin, et al. "ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA." Magnetic resonance in medicine 71.3 (2014): 990-1001.
8. Zbontar, J., Knoll, F., Sriram, A., Murrell, T., Huang, Z., Muckley, M. J., ... & Lui, Y. W. (2018). fastMRI: An open dataset and benchmarks for accelerated MRI. arXiv preprint arXiv:1811.08839.
9. Moeller, Steen, et al. "Multiband multislice GE‐EPI at 7 tesla, with 16‐fold acceleration using partial parallel imaging with application to high spatial and temporal whole‐brain fMRI." Magnetic resonance in medicine 63.5 (2010): 1144-1153.
Figures