0948
Optimizing the NEXI acquisition protocol for quantifying human gray matter microstructure on a clinical MRI scanner using Explainable AI1Dept. of Radiology, Lausanne University Hospital (CHUV), Lausanne, Switzerland, 2Siemens Healthcare GmbH, Erlangen, Germany, 3Signal Processing Lab 5 (LTS5), École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
Synopsis
Keywords: Microstructure, Modelling, Acquisition Protocol
We optimized the acquisition protocol for parameter estimation of the NEXI model, suited to characterize gray matter microstructure, using concepts from Explainable AI. The improvement over a “naïve” protocol was only marginal. The limit on NEXI parameter estimation precision and accuracy is largely driven by the model and the type of measurements available (linear diffusion encoding, in a combination of (b,t) pairs) and is likely already reached as can be estimated from CRLB. The silver lining is that a clinical acquisition protocol feasible on a 3T system with 80 mT/m gradients yields reasonable NEXI microstructure maps in the human brain.
Background
NEXI1 (or SMEX2) is a two-compartment diffusion model that accounts for inter-compartment exchange. It is suited to characterize gray matter (GM) microstructure; its parameters are the inter-compartment exchange time tex, the intra and extra-neurite apparent diffusivities Di and De and the intra-neurite signal fraction f. It has first been implemented in a preclinical setting in vivo1 and ex vivo2. Here, we aim to optimize the acquisition protocol for NEXI parameter estimation and demonstrate feasibility on a clinical 3T system.Methods
Simulated data: Synthetic signals were generated using the NEXI model for a large number of parameter combinations, with Rician noise and SNR at b=0 ranging 80 – 120, assuming 3 different acquisition protocols. Parameter combinations included tex ranging [0 – 150]ms, Di and De [0.1 – 3.5]µm²/ms and f [0.1 – 0.9]. Each protocol sets a total number of measurements N and the associated b-value and diffusion time (b,t) pairs. For each protocol, one multi-layer perceptron (MLP) of 3 hidden layers and 500 neurons per layer was trained on 2.106 random sets and applied to a test set of 104 other examples, taking as input the simulated signals and as output the corresponding NEXI parameter combinations. The first ‘Extended’ protocol was a grid of 14 b-values (1 – 7.5ms/µm2) and 12 diffusion times (20 – 47.5ms) (N=168). The Shapley values of this network were calculated on a subset of training and test sets using SHAP3 to estimate the importance of each (b,t) pair in the NEXI parameter estimations. The (b,t) pairs with highest importance were used to establish an ‘Optimized’ protocol with a realistic number of measurements (N=16). This protocol was compared to a ‘Clinical’ grid-like protocol, based on the mean absolute error (MAE) of each parameter estimation derived from these protocols. Cramer Rao Lower Bounds (CRLB) of both protocols were finally computed from simulated signals obtained from several ranges of Ground Truths. Clinical data: The study was approved by the relevant ethics board. One male healthy volunteer was scanned. Acquisition: Diffusion-weighted images were acquired on a 3T system with 80 mT/m gradients (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany), using a PGSE-EPI research sequence with the following parameters: b-values=1.00, 2.00, 3.20, 4.44 and 5.00ms/µm², 20 directions per shell, diffusion times Δ=28.3, 36.0, 45.0, 55.0 and 65.0ms, δ=16.5ms, 4 b=0 images, spatial resolution=2x2x2 mm3, total scan time: 35min. The (b,t) pairs corresponded to the “Clinical” protocol. Processing: Multi-shell multi-diffusion time data were preprocessed jointly; steps included MP-PCA magnitude denoising4, Gibbs ringing5, distortion and Eddy current corrections6. Parametric maps of the NEXI model parameters were estimated from the powder-average signals using non-linear least-squares (NLS) and the MLP network trained previously on the Clinical protocol.Results and discussion
The most important (b,t) pairs for each of the NEXI parameters vary (Fig. 1): (low b, low t) and (high b, high t) combinations for tex, low b and full-range of t for Di, low-medium b and medium-long t for both De and f. To produce an optimized protocol of N=16 (b,t) pairs, we ranked for each parameter the best (b,t) pairs by mean absolute SHAP value, weighted by the sign of the median SHAP value. Based on the RMSE on each parameter in the Extended MLP, normalized by the parameter range, we assigned 8 optimal pairs for tex, 4 for Di, 2 for De and 2 for f (Fig. 2). In other words, the parameter with highest normalized RMSE got the largest number of optimal (b,t) pairs for its estimation. Remarkably, the MAE of the MLPs trained on the Optimized protocol was only slightly lower than that of the Clinical protocol (Table 1). The Optimized protocol also yielded slightly lower CRLB (Fig.3) for typical experimental NEXI values (see Fig.4), which suggests the variances of experimental estimates could potentially be further reduced. The marginal improvement does not however allow us to favor one protocol over the other, especially as some pairs of the Optimized protocol are difficult to achieve on a clinical system. Using the Clinical protocol, we report the first microstructure maps of the NEXI model estimated in the human brain on a conventional clinical scanner, using NLS and MLP (Fig.4). NLS results in numerous voxels hitting the permitted parameter bounds, while MLP does less. Using a cortical ribbon mask, we estimate typical NEXI parameter values in the cortex, which are consistent between NLS and MLP, as well as with previous estimates in the rat cortex1.Conclusion
The limit on NEXI parameter estimation precision and accuracy is largely driven by the model and the type of measurements available (linear diffusion encoding, in a combination of (b,t) pairs) and only leaves room for moderate improvement as can be estimated from CRLB. The silver lining is that a clinical acquisition protocol feasible on a 3T system with 80 mT/m gradients yields reasonable NEXI microstructure maps in the human brain. Future work will focus on estimating the robustness of these maps across subjects, as well as search for improved strategies in denoising and possibly data coverage (e.g. multi-dimensional MRI measurements) to lower the CRLB’s nonetheless. The effect of ‘fat’ diffusion pulses will also be investigated, as suggested previously2.Acknowledgements
QU, TP and IJ are supported by SNSF Eccellenza grant PCEFP2_194260. EJC-R was supported by SNSF Ambizione grant PZ00P2_185814.References
[1] Jelescu, Neuroimage 2022 [2] Olesen, NeuroImage 2022 [3] Lundberg and Lee, NIPS 2017 [4] Veraart, NeuroImage 2016 [5] Kellner, MRM 2016 [6] Andersson, NeuroImage 2016.Figures
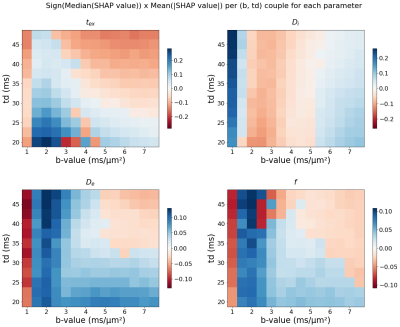
Figure 1. Mean absolute SHAP values of each NEXI parameter for each (b,t) pair in the Extended protocol, weighted by the sign of the median SHAP values. The signs allow to extract two different regions of high importance for tex estimation.
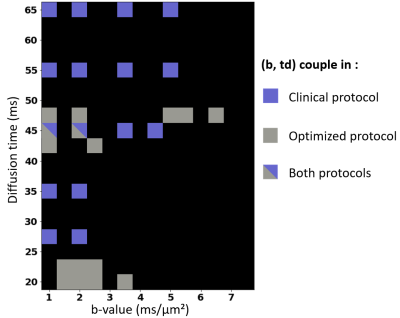
Figure 2. (b,t) pairs selected in the Clinical and Optimized protocols.
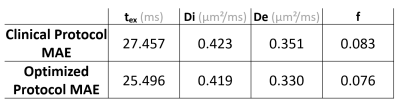
Table 1. Mean absolute error of the MLP with each input protocol
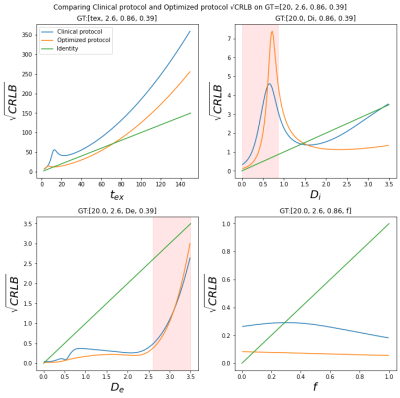
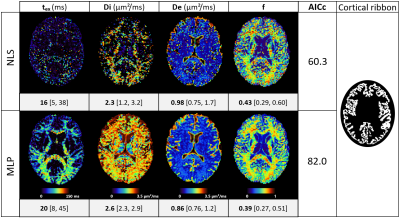
Figure 4. Axial slice of NEXI parametric maps in one subject using Non-Linear Least Squares (NLS) or the trained Multi-Layer Perceptron (MLP) and Akaike information criterion (AICc) of each method. Median values and quartiles of the parameter estimations refer to the cortical ribbon, shown on the right. Only voxels where the fit converged were included in the statistics (9% of mask voxels for NLS, and 58% for MLP). MLP estimates are very different from the mean of the training prior.