0947
Deep-Learning Enhancement of 3D-MSI using Conventional Images as Training Targets1Radiology, Medical College of Wisconsin, Milwaukee, WI, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Image Reconstruction
We present a feasibility study exploring the use deep learning (DL) methods to enhance the image quality of isotropic 3D-multi-spectral metal artifact suppressed images. Conventional high resolution 2D fast/turbo spin echo images are used as network training labels by masking regions corrupted by metal artifacts. A pilot set of 3 cases deploying the presented concept to T2-weighted imaging of the instrumented spine are analyzed. The quantitative results of this analysis demonstrate improved spinal cord contrast, image resolution, and general agreement with 2D-FSE images when the DL enhancement model is inferred on the complete 3D-MSI datasets.Introduction
3D Multi-Spectral Imaging (3D-MSI) methods [1,2,3] are capable of substantially reducing artifacts from metal implants located in the vicinity of areas of clinical assessment. Though artifact suppression using 3D-MSI is sufficient for clinical diagnostics, the resolution and contrast of 3D-MSI is well-below the capabilities of conventional 2D fast/turbo spin echo (2D-FSE) sequences. These limitations are due to a variety of factors that enable 3D-MSI to suppress artifacts.In the present study, we seek to improve the quality of isotropic 3D-MSI acquisitions [4], which have superior through-plane resolution, but lack the in-plane resolution of conventional 2D-FSE images. The basic premise of our image enhancement approach leverages the 2D-FSE images that are typically acquired in addition to 3D-MSI, which provide higher quality assessments in regions not contaminated by metal artifacts. By automatically identifying regions where the 2D-FSE image are not contaminated, they can be used as a deep-learning (DL) training label to construct inferencing models to globally improve the quality of 3D-MSI.
Methods
Deep-Learning Image Enhancement Approach:The key concept to our approach for 2D-FSE-informed enhancement of 3D-MSI requires identification of regions where the 2D-FSE can be used as targets (or labels) of desired 3D-MSI contrast. This requires 2D-FSE and 3D-MSI images of similar contrast, where the 2D-FSE resolution and contrast are more desirable than that of the 3D-MSI.
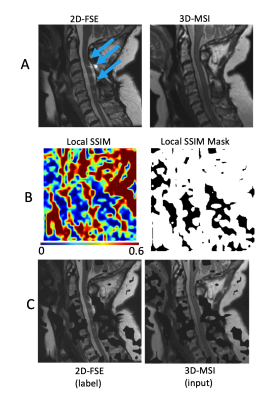
After resampling and registration of 3D-MSI to the 2D-FSE reference images, a local structural similarity image metric (SSIM) map is constructed. This is accomplished by downsampling both image pairs to a common resolution and computing SSIMs across overlapping patches. 32x32 patches over a 512x512 in-plane target matrix (with 50% overlap) were utilized for the present demonstration. This local SSIM map was thresholded to create a mask of regions where the two images nominally matched one another at a low resolution baseline.
A 2D convolutional neural net (CNN) was repurposed from an open-source super-resolution concept repository [4]. The only customizations made to this CNN were a) the removal of upscaling and subpixel convolutions, b) the use of Parametric Rectified Linear Unit (PReLU) activations, 64 latent channels, and a kernel size of 5. The adam optimizer was used with a mean-squared error loss function across 500 epochs. Training and inferencing were performed on a NVIDIA Titan B with 12GB of memory.
Evaluation Subjects:
This feasibility study was performed in order improve the quality of 3D-MSI being utilized within a broader study of instrumented treatment of spinal cord degeneration and injury. For the present pilot analysis, data from 3 subjects with instrumented spinal fusion treatment of degenerative cervical myelopathy were analyzed. These subjects provided written consent into an a locally IRB protocol.
MRI Data:
T2-weighted 3D-MSI were acquired with 1.2 mm isotropic resolution with TE=60ms and TR = 2.5s. Sagittal 2D-FSE images (TE = 106ms, TR = 3s) were acquired with 15 3 mm slices and 0.4x0.6 mm in-plane resolution, which was zero-filled to 0.3x0.3 resolution by the vendor reconstruction. 3D-MSI were resampled to the 2D-FSE spatial domain using Advanced Normalization Tools (ANTs) software.
Analysis: After training and inferencing of the enhanced 3D-MSI, SSIM of the masked input and predicted slices were computed against the masked 2D-FSE training slices. In addition, several demonstrative profiles were extracted from the images to examine the spinal cord contrast, image resolution, and spinal cord lesion conspicuity enabled by the enhancement process.
Results
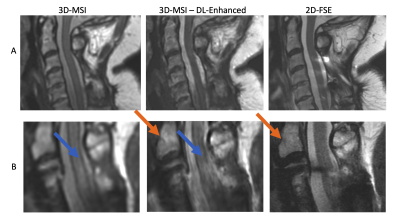
Figure 1 provides representative images and maps outlining the data curation process. Row c) provides exemplary images that were input into the CNN training algorithm.Figure 2 demonstrates the capabilities of this preliminary enhancement concept. The cord contrast and image resolution improvements are clearly evident in the displayed images. Row B) demonstrates the potential clinical utility of this enhancement algorithm, where a cord hyperintensity lesion is completely obscured by artifact in the 2D-FSE images, only mildly visible in the sub-par contrast of the original 3D-MSI, but is sharply visible in the enhanced 3D-MSI.
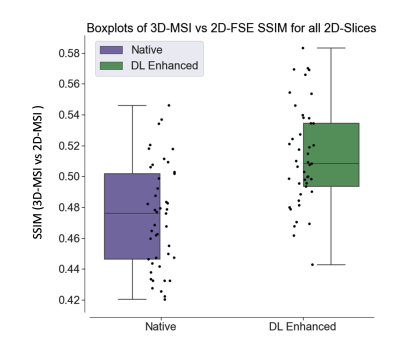
Figure 3 provides graphical evidence of the SSIM improvements gained by the enhancement algorithm against the target 2D-FSE images. The scale of the SSIM is not high, but the enhancement process makes a highly statistically significant (p<0.0001) advance in moving the 3D-MSI closer to the 2D-FSE image target. The measured SSIM improvement across the test data was 8.3 +/3.0 %.
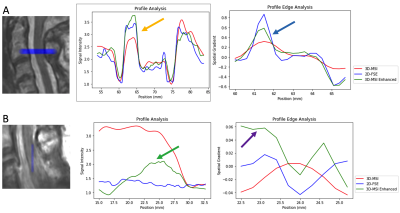
Figure 4 utilizes image trace profiles (indicated in image overlays) to illustrate the impact of the 3D-MSI enhancements on cord contrast, resolution, and cord lesion conspicuity.
Discussion
This study has demonstrated a promising feasibility of utilizing 2D-FSE images as label targets for deep-learning enhancement of 3D-MSI. The challenges posed by suppressing metal artifacts are substantial and 3D-MSI struggles to match the quality of imaging methods that do not attempt to suppress these artifacts. The presented methods offer a solution whereby conventional images, which provide superior image quality away from metal implants, can be used to improve the image quality of 3D-MSI near metal interfaces.This technical demonstration was preliminary in nature. A multitude of improvements to the training approach and CNN design will be explored in future work. In addition, generalized networks that can enhanced 3D-MSI without a paired training set will also be explored.
Acknowledgements
This work was partially supported by the Department of Defense Congressionally Directed MedicalResearch Program, Spinal Cord Injury Research Program, award number W81XWH1910273. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense This research was also supported by National Institute of Health (NIH) grant R21EB030123.References
[1] Koch KM, Lorbiecki JE, Hinks RS, King KF. A multispectral three-dimensional acquisition technique for imaging near metal implants. Magn Reson Med. 2009 Feb;61(2):381-90.
[2] Lu, W., Pauly, K. B., Gold, G. E., Pauly, J. M., & Hargreaves, B. A.. SEMAC: slice encoding for metal artifact correction in MRI. Mag Reson Med,2009, 62(1), 66-76.
[3] Koch KM, Brau AC, Chen W, Gold GE, Hargreaves BA, Koff M, McKinnon GC, Potter HG, King KF. Imaging near metal with a MAVRIC-SEMAC hybrid. Magn Reson Med. 2011 Jan;65(1):71-82.
[4] https://github.com/sgrvinod/a-PyTorch-Tutorial-to-Super-Resolution
[5] He, K., Zhang, X., Ren, S., & Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the IEEE international conference on computer vision. 2015. (pp. 1026-1034).
Figures