0945
Prediction of MGMT Methylation Status of Gliomas Using Pre-operative MR Images: a Fully Automatic Convolutional Neural Networks Based Approach
Xiaohua Chen1, Zhiqiang Chen2, Zhuo Wang1, Shaoru Zhang1, Yunshu Zhou1, Shili Liu1, Ruodi Zhang1, Yuhui Xiong3, and Aijun Wang4
1Clinical medicine school of Ningxia Medical University, Yinchuan, China, 2Department of Radiology ,the First Hospital Affiliated to Hainan Medical College, Haikou, China, 3GE Healthcare MR Research, Beijing, China, 4Department of Radiology, General Hospital of Ningxia Medical University, Yinchuan, China
1Clinical medicine school of Ningxia Medical University, Yinchuan, China, 2Department of Radiology ,the First Hospital Affiliated to Hainan Medical College, Haikou, China, 3GE Healthcare MR Research, Beijing, China, 4Department of Radiology, General Hospital of Ningxia Medical University, Yinchuan, China
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Brain
This study aims to propose a fully automatic approach based on convolutional neural networks (CNNs) to predict the O6-Methylguanine-DNA-methyltransferase (MGMT) methylation status of gliomas using conventional pre-operative MR images. It was shown that the Markov Random Field-U-Net network can accurately segment the tumor region, and the improved 34-layer Resnet network can predict the MGMT methylation status effectively. This model has the potential to be a practical tool for the non-invasive characterization of gliomas to help the individualized treatment planning.Summary of Main Findings
A deep learning model was proposed that can automatically predict the MGMT methylation status of gliomas using pre-operative MR images.Synopsis
This study aims to propose a fully automatic approach based on convolutional neural networks (CNNs) to predict the O6-Methylguanine-DNA-methyltransferase (MGMT) methylation status of gliomas using conventional pre-operative MR images. It was shown that the Markov Random Field-U-Net network can accurately segment the tumor region, and the improved 34-layer Resnet network can predict the MGMT methylation status effectively. This model has the potential to be a practical tool for the non-invasive characterization of gliomas to help the individualized treatment planning.Introduction
Glioma is the most common malignant primary brain tumor in adults, and it is a highly heterogeneous disease with various molecular subtypes and different treatment strategies or clinical prognosis1, 2. O6-Methylguanine-DNA-methyltransferase (MGMT) promoter methylation confers an improved prognosis and treatment response in gliomas. Thus, determining MGMT promoter methylation status is important in predicting survival rate or designing treatment plan3, but there is no reliable and non-invasive way to achieve it. Therefore, considerable attention has been dedicated to developing image-based diagnostic methods to determine MGMT promoter methylation status. Convolutional neural network (CNN) is a representative method to exploit high-dimensional numeric information from images by learning relevant features directly from image signal intensities, and it is being studied in great demand in glioma molecular classification. The purpose of the study was to predict the MGMT promoter methylation status of patients with gliomas (grades II-IV) from pre-operative MR images using an automatic approach that integrates (i) CNN-based tumor segmentation and (ii) CNN-based MGMT status prediction. The model shows in Figure 1.Methods
170 patients (105 male,50.6±3.9years,65 male,47.3±1.3years) were retrospectively included in our study. The inclusion criteria were as follows: (i) pathologically confirmed glioma, (ii) known MGMT status, (iii) preoperative MRI inclusive of CE-T1WI, T2WI ,T1WI,T2flair, and (iv) age ≥18 years. All patients underwent MR exams on a 3.0 T MR scanner (SIGNATM Architect; GE Healthcare, Milwaukee, WI, USA) with a 48-channel head coil. The scan protocol and detailed parameters were listed in Table 1. Our automatic process includes 2 models. Model 1 is a network that combines Markov Random Field (MRF) with U-Net, which was used to segment tumor into edema, hemorrhage and necrosis in tumor. Details showed in Figure 2.The images were standardized and signal intensity normalized firstly. Then the images were randomly divided into training set, verification set and test set according to the ratio of 6:2:2. The training set images with sizes of 512×512×1 as network parameters of the model. Our CNN classifier for MGMT status prediction (model 2) is derived from the well-known 34-layer Resnet architecture (hereinafter referred as the conventional G-Resnet34). Improvements made on this architecture were shown in Figure 2. The model images input comprised tumor masks of 512 ×512size and axial CE-T1WI, T2WI, T2-FLAIR and T1WI images. The performance of Model 1 for tumor segmentation was measured using the dice similarity coefficient (Dice), PPV (positive predictive value), sensitivity. The diagnostic performance of model 2 was measured in terms of accuracy, area under the receiver operating characteristic curve (AUC) and F1. The 10-fold cross-validation was used to verify the stability of model 2.Results
All parameters were calculated in PyCharm using Python. Model 1 (CNN for tumor segmentation) yielded dice coefficients of 0.976 averagely. The PPV and sensitivity were 0.947 and 96.8%. The segmentation result was shown in Figure 3. Our model 2 which predicted the MGMT status achieved accuracies of 99.3% (AUC = 0.96),96.5% (AUC = 0.91),97.3% (AUC = 0.96), with F1 of 98.6%,98.6%,95.9%in the training set, verification set and test set, respectively (Figure 5). The average accuracy of 10-fold cross validation was 95.8%.Discussion and Conclusion
An MGMT promoter methylation status confers a better prognosis and treatment response of gliomas, independent of the histologic grade3. Our study demonstrated that the automatic approach based on conventional MRI images can accomplish the task from glioma tumor segmentation to the prediction of MGMT promoter methylation. Time-consuming and subjective differences can be avoided because of automatic segmentation of tumor regions. Our model of segmentation derived from U-Nets, and was combined with MRF to extract the hierarchical features of the spatial location of the tumor region to improve the segmentation accuracy further. Besides, the gaussian filter on both sides of the network to refine the tumor area, so as to improve accuracy of the results. We build the prediction model based on Resnet34. Besides, we combined the first and the third layer, the second and the fourth layer of the network. So as to fuse the deep and shallow layers better, which allows obtain more features to improve the accuracy of the model. In addition, a large amount of data is required to deep learning model that ensure the stability and prevent over fitting. One of the major limitations in our study is the relatively small number of lesions. Hence future study with bigger cohort of subject is warranted. To conclude, we can predict the MGMT methylation status of gliomas using a fully automatic CNNs based on conventional MR imaging.Acknowledgements
No acknowledgement found.References
[1] MOLINARO A M, TAYLOR J W, WIENCKE J K, et al. Genetic and molecular epidemiology of adult diffuse glioma [J]. Nat Rev Neurol,2019:405-17.
[2] LOUIS D N, PERRY A, WESSELING P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary [J]. Neuro-Oncology,2021:1231-51.
[3] ELLINGSON B M, CLOUGHESY T F, POPE W B, et al. Anatomic localization of O6-methylguanine DNA methyltransferase (MGMT) promoter methylated and unmethylated tumors: a radiographic study in 358 de novo human glioblastomas [J]. Neuroimage,2012:908-16.
Figures
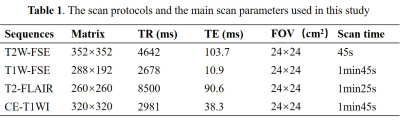
Abbreviation: T2W-FSE: T2-weighted fast spin
echo; T1W-FSE: T1-weighted fast spin echo; T2-FLAIR: T2-weighted fluid attenuation inversion recovery; CE-T1WI: contrast-enhanced
T1-weighted .Slice
thickness/gap = 5/1mm; all images were collected in transverse view.
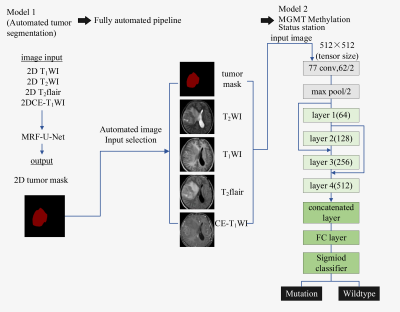
Fig. 1 Fully automated hybrid model for MGMT methylation status prediction.
In Model 2, layers 1–4 consisted of 3,4,6, and 3 residual blocks, with each
block containing the 3 ×3convolutions twice. Fused the first and the third
layer, the second and the fourth layer of the network structure.
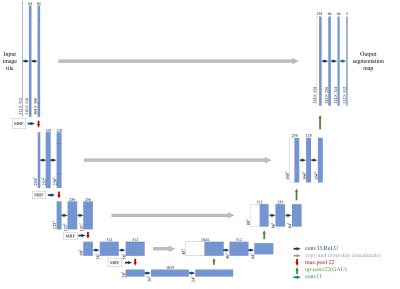
Fig. 2 MRF-U-Net
Architecture Overview. Combining MRF and U-Net to segment the tumor of gliomas.
The MRF were localized between the down sampling layers.
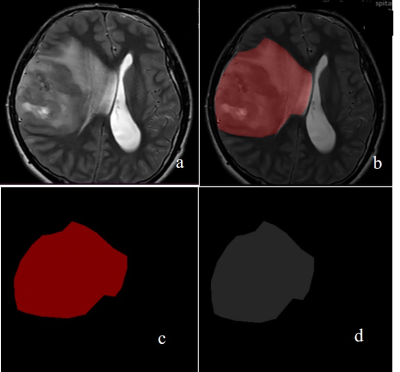
Fig. 3: The segmentation results of model 1. (a) original
image of T2WI; (b) labelled image; (c) tumor mask; (d)segmentation module.
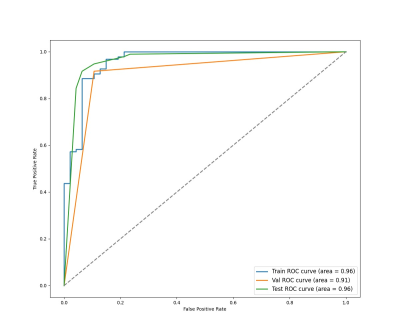
Fig. 4: The ROC
curves for the G-ResNet34 to predict the MGMT methylation status of gliomas.
DOI: https://doi.org/10.58530/2023/0945