0931
Deep learning based segmentation of aortic cross sections (2D+t) in multi-vendor 4D PCMRI1Institute of Computer-assisted Cardiovascular Medicine, Charité – Universitätsmedizin Berlin, Berlin, Germany, 2Fraunhofer MEVIS, Bremen, Germany, 3IBM Germany, Berlin, Germany, 4DZHK (German Center for Cardiovascular Research), Partner site Berlin, Germany, 5Charité – Universitätsmedizin Berlin, Berlin, Germany, 6University Hospital Freiburg, Freiburg, Germany, 7University Medical Center Hamburg-Eppendorf, Hamburg, Germany
Synopsis
Keywords: Flow, Velocity & Flow, Aorta segmentation
Standardized 4D PCMRI postprocessing protocols could enable comparable bloodflow quantification. We propose an automatic segmentation of aortic cross section over time with a residual trained data from different imaging sequences, scanner types, pathologies and position of cross section planes. Dice score, Hausdorff metric as well as flow and velocity curves for the segmented areas show good performance both in the validation and test sets.Introduction
4D PCMRI enables comprehensive assessment of aortic bloodflow, and medical societies suggest standardized postprocessing protocols to enable comparative quantitative analysis. Data acquired with different protocols, scanners and sequences can differ regarding intensity ranges and velocity encoding. Existing approaches for 3D and 4D segmentation of 4D PCMRI data achieve good results for specific imaging sequences and scanner manufacturers [1-3]. The authors propose a multi-site ML model for automatic segmentation of aorta cross sections over time and evaluate the results in terms of accuracy of segmentation masks and the effect of segmentation differences on derived clinical parameters such as throughflow.Methods
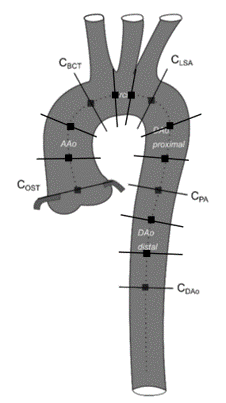
We considered 4D-flow MRI data from 205 subjects; 130 without known aortic pathology (Siemens Trio Tim 3T) [4], 59 with aortic stenosis (Philips Achieva 1.5T) [5], and 8 healthy volunteers (Siemens Prisma Fit 3T, Philips Ingenia 3T) [6]. Per dataset 11-12 planes (Figure 1) were placed perpendicular to the aortic centerline obtaining a total number of 2316 cross sections, and lumen boundaries were annotated by medical experts (21 to 57 frames/cardiac cycle depending on the heart rate).A residual Unet [7] was trained to generate 3D aortic cross sectional segmentations (x,y,t) on preprocessed (phase unwrapping, offset correction) 4D flow MRI data. 2D+t planes of magnitude and velocities fields were prepared for processing as follows:
- Z-score normalization
- Spatiotemporal resampling to obtain a final in-plane resolution of 1.54 mm2 and 64 timesteps
- Padding/cropping to an in-plane dimension of 64x64
- Concatenation of magnitude and velocity fields as channels.
$$v(t) = \frac{1}{\mid A(t)\mid} \int_{A(t)}^{}\parallel v(x,y,t)\parallel dA$$
$$I(t) = \int_{A(t)}^{} <v(x,y,t),n>dA$$
With:
- ||v|| the magnitude of the velocity vector
- v(x,y,t) the velocity vector in the point (x,y) at time t
- |A(t)| the area of the segmentation on timeframe t
- n normal vector of the cross-sectional plane
- And <a,b> denotes the scalar product between the vectors a and b
Results
The data were randomly divided into 88% for training, 10% for validation and 2% for testing (see Figure 1). DS and HD were:- Validation set: DS = 0.89±0.04, HD = 2.99±1.89
- Test set: DS = 0.89±0.12, HD = 3.95±2.17.
Discussion
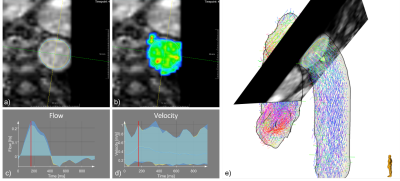
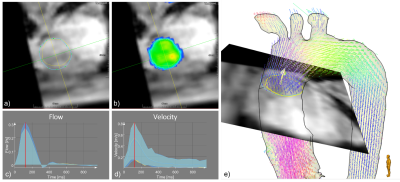
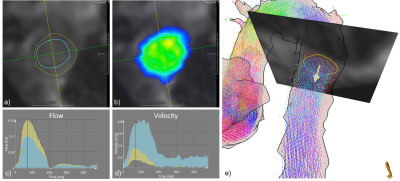
The 3D Unet trained on the multi-scanner, multi-sequence dataset showed good performance for the different manufacturers, pathologies, and cross section positions for all age groups (21-81). The automatic segmentation provided flow and velocity curves comparable with expert analyses. The inspection of cases with lower DS indicates that the model might perform better than the expert in low contrast regions (Figure 4).Conclusion
With a multicenter, multiscanner, multisequence dataset it might be possible to provide automatized segmentation methods, which enable comparable measurements on 4D flow datasets acquired in different scenarios.Acknowledgements
Funding from the German Research Foundation (GRK2260, BIOQIC).
Funding by the German Research Foundation (DFG) as part of SFB-1470, B06.
References
[1] Berhane H, Scott M, Elbaz M, Jarvis K, McCarthy P, Carr J, Malaisrie C, Avery R, Barker AJ, Robinson JD, Rigsby CK, Markl M. Fully automated 3D aortic segmentation of 4D flow MRI for hemodynamic analysis using deep learning. Magn Reson Med. 2020 Oct;84(4):2204-2218. doi: 10.1002/mrm.28257.
[2] Pradella M, Scott MB, Omer M, Hill SK, Lockhart L, Yi X, Amir-Khalili A, Sojoudi A, Allen BD, Avery R, Markl M. Fully-automated deep learning-based flow quantification of 2D CINE phase contrast MRI. Eur Radiol. 2022 Oct 29. doi: 10.1007/s00330-022-09179-3.
[3] Bustamante, M., Viola, F., Engvall, J., Carlhäll, C.-J. and Ebbers, T. (2022), Automatic Time-Resolved Cardiovascular Segmentation of 4D Flow MRI Using Deep Learning. J Magn Reson Imaging. https://doi.org/10.1002/jmri.28221
[4] Harloff A, Hagenlocher P, Lodemann T, Hennemuth A, Weiller C, Hennig J, Vach W. Retrograde aortic blood flow as a mechanism of stroke: MR evaluation of the prevalence in a population-based study. Eur Radiol. 2019 Oct;29(10):5172-5179. doi: 10.1007/s00330-019-06104-z.
[5] Nordmeyer S, Lee CB, Goubergrits L, Knosalla C, Berger F, Falk V, Ghorbani N, Hireche-Chikaoui H, Zhu M, Kelle S, Kuehne T, Kelm M. Circulatory efficiency in patients with severe aortic valve stenosis before and after aortic valve replacement. J Cardiovasc Magn Reson. 2021 Mar 1;23(1):15. doi: 10.1186/s12968-020-00686-0. PMID: 33641670; PMCID: PMC7919094.
[6] Demir A, Wiesemann S, Erley J, Schmitter S, Trauzeddel RF, Pieske B, Hansmann J, Kelle S, Schulz-Menger J. Traveling Volunteers: A Multi-Vendor, Multi-Center Study on Reproducibility and Comparability of 4D Flow Derived Aortic Hemodynamics in Cardiovascular Magnetic Resonance. J Magn Reson Imaging. 2022 Jan;55(1):211-222. doi: 10.1002/jmri.27804. Epub 2021 Jun 25. PMID: 34173297.
[7] Anita Khanna, Narendra D. Londhe, S. Gupta, Ashish Semwal, A deep Residual U-Net convolutional neural network for automated lung segmentation in computed tomography images, Biocybernetics and Biomedical Engineering,Volume 40, Issue 3,2020,Pages 1314-1327,ISSN 0208-5216, https://doi.org/10.1016/j.bbe.2020.07.007.
Figures