0897
Deep Learning Based Automated Multi-Organ Segmentation in Lymphoma Patients using Whole Body Multiparametric MRI Images
Anum Masood1,2, Sølvi Knapstad2, Håkon Johansen3, Trine Husby3, Live Eikenes 1, Pål Erik Goa2,3, and Mattijs Elschot 1,3
1Department of Circulation and Medical Imaging, NTNU, Trondheim, Norway, 2Department of Physics, NTNU, Trondheim, Norway, 3Department of Radiology and Nuclear Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway
1Department of Circulation and Medical Imaging, NTNU, Trondheim, Norway, 2Department of Physics, NTNU, Trondheim, Norway, 3Department of Radiology and Nuclear Medicine, St. Olavs Hospital, Trondheim University Hospital, Trondheim, Norway
Synopsis
Keywords: Segmentation, Data Processing, Automated Segmentation, Deep Learning, nnUnet
Widespread of lymphoma cancer makes manual segmentation of metastatic lymph nodes a tedious task. Lymphoma cancer is assigned an anatomic stage using the Ann Arbor system which relies on the segmentation and localization of affected lymph nodes with respect to anatomical stations. We present a framework for multi-organ segmentation for multiparametric MRI images. Our modified nnUnet using a transfer learning approach achieved 0.8313 mean DSC and 0.659 IoU in lymphoma cancer dataset.Introduction
Lymphoma cancer develops in the lymphatic system and can affect organs throughout the body. Lymphoma cancer is assigned an anatomic stage using the Ann Arbor system which relies on segmentation and localization of affected lymph nodes. As lymphoma Ann-Arbor Staging is based on lymph nodes localization above or below the diaphragm, we propose using the location of nearby organs as prior information to guide the lymphoma stage classification. In order to automate lymphoma cancer detection and staging, the location of the lesion with respect to these stations has to be determined. Widespread lymphoma makes manual segmentation of metastatic lymph nodes a tedious task. We aim to develop a method for automated segmentation and localization of lymph nodes.Method
We present a framework for multi-organ segmentation for multiparametric MRI images. Our model is based on nnU-Net, a validated model for multi-organ segmentation using CT images. We used the transfer learning technique and initially trained our model on the dataset provided by nnU-Net developers 1. Furthermore, we used an in-house dataset comprised of multiparametric MRI images of 22 lymphoma patients, with manual segmentations of multiple anatomical landmarks (heart, left lung, right lung, liver, spleen, urinary bladder, pelvic bone, abdomen, left kidney, right kidney, teeth, cervical vertebrae, aorta, and mediastinum) validated by a radiologist. Initially, we trained nnU-Net with no change to provide a baseline for further modifications using T2 Haste MRI, T2 TIRM MRI, and Diffusion Weighted Image (DWI) with b = 800 s/mm2. nnU‐Net automatically self-configures and runs the entire segmentation process including pre-processing, data-augmentation, training, and post-processing steps. Secondly, we ran the nnU-Net with some modifications. For our training model, we used binary cross-entropy instead of using cross-entropy loss which optimized each of the regions independently. In addition to the nnU-Net original data-augmentation methods, we increased scaling range, rotation probability, and brightness augmentation for enhanced data augmentation. We also increased the batch size from 2 to 3 for improving model accuracy, but due to the relatively smaller dataset, this resulted in overfitting. All experiments were run as five-fold cross-validation.Results & Discussion
We carried out experiments for 14 organ segmentation, and compared results with the ground truth, leading to promising results, presented in Figure 2 and Figure 3. We aimed to investigate the accuracy of the computer-aided segmentation using the mean Dice Similarity Coefficient (DSC), mean Intersection over Union (IoU), Precision, Recall, and F1 Score performance metric for the evaluation of 14 organ delineations in the testing cohort. nnU-Net achieved 0.5145 mean DSCs and 0.3162 IoU for our initial dataset whereas the nnU-Net model with modifications using a transfer learning approach achieved 0.8313 mean DSC and 0.659 IoU. Owing to the small cohort, the relatively low DSCs value of 0.8313 was observed in our experiment. These preliminary results highlight the potential of nnU-Net for automated multi-organ segmentation.Conclusion
Using a large dataset with a wide range of variations in terms of lymph nodes' location, shape, or contrast uptake will be used for further evaluation. The automated segmented organs could potentially be used in the next step to assign affected lymph nodes to their respective stations, thereby creating a fully automated system for Ann-Arbor staging.Acknowledgements
This work was supported by the Trond Mohn Foundation for 180°N (Norwegian Nuclear Medicine Consortium) project.References
[1] Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J., & Maier-Hein, K. H. (2021). nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2), 203-211.Figures
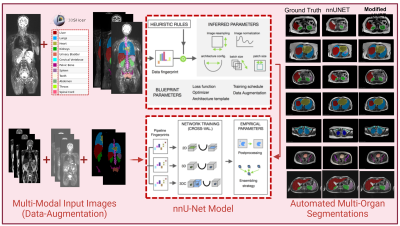
Figure 1:
Overview of Automated Multi-Organ Segmentation using Modified nnU-Net. Multiparametric
MRI images and manual segmentations are used as input and modified nnU-Net predicts multi-organ segmentations. Segmentation results of nnUnet and modified nnU-Net are compared with the ground truth.
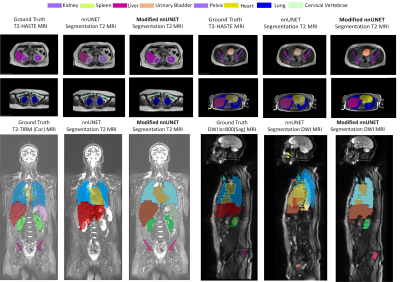
Figure 2: Visual comparison of segmentation results of the modified nnUnet with state-of-the-art nnUnet on multi-modal MRI Lymphoma Dataset. First two rows consist of a raw T2 HASTE (Axial View) MRI image with ground truth, prediction by nnU-NET, and segmentation predicted by modified nnU-Net. The third row consists of two sets of results, comparisons of predicted segmentation by nnUnet and modified nnUnet against ground truth using T2
TIRM (Coronal View) MRI and DWI (Sagittal View).
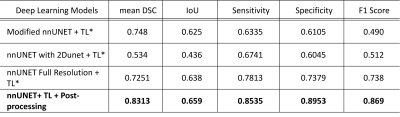
Figure 3: Quantitative
results using performance metric; mean Dice Similarity Coefficient (DSC), mean
Intersection over Union (IoU), Precision, Recall, and F1 Score for the automated
organ segmentation using modified nnU-NET.
DOI: https://doi.org/10.58530/2023/0897