0891
Anatomical MRI-guided deep learning-based low-count PET image recovery without the need for training data – a PET/MR study1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Psychiatry, Stony Brook Medicine, Stony Brook, NY, United States, 3Radiology, Stony Brook Medicine, Stony Brook, NY, United States
Synopsis
Keywords: PET/MR, PET/MR
The advent of simultaneous PET/MRI enables the possibility of using MRI to guide PET image reconstruction/recovery. Deep-learning approaches have been explored in low-count PET recovery, with current approaches focus on supervised learning, which requires a large amount of training data. A recently proposed unsupervised learning image-recovery approach does not require this but relies on the optimal stopping criterion. In this work, we developed an unsupervised learning-based PET image recovery approach using anatomical MRI as input and a novel stopping criterion. Our method achieved better image recovery in both global image similarity metrics and regional standard uptake value (SUV) accuracy.
Introduction
Low-count PET is not only capable of reducing the injected dose, but also can be used to reduce acquisition time. However, the image quality of low-count PET can be too low for clinical use if reconstructed using conventional reconstruction approaches. To solve this, deep learning techniques have been proposed to yield high-count-like images from low-count. Most current techniques utilize supervised learning, where a large amount of high-count and low-count PET image pairs are needed for the network to learn. However, this can be difficult to obtain especially for newly developed tracers, and could potentially be disease-dependent.Unsupervised learning has been proposed for this task utilizing anatomical MRI images. This approach does not require paired low-count and high-count data. Unsupervised learning relies on the optimal stopping criteria to stop the training at the desired point. A recent study used contract-to-noise ratio (CNR) as the stopping criterion for tumor contrast recovery1. In this work, we proposed an improved stopping criterion for this novel approach that can be used for general brain studies with improved global image similarity and accurate regional uptake values.
Methods
PET/MRI dataA total of 28 18F-FDG brain PET/MRI studies were gathered from our previously acquired research studies. None of these patients had brain tumors or neurological diseases. All data sets used listmode acquisition with a 20-channel PET-compatible MRI head coil on a Siemens Biograph mMR PET/MRI scanner. T1-weight MRI images were acquired using MPRAGE sequence (TE/TR/TI = 3.24/2300/900 ms, flip angle = 9, voxel size = 0.87$$$\times$$$0.87$$$\times$$$0.87mm3). Attenuation maps were generated using an established MPRAGE-based algorithm which incorporates bone compartment2,3.
PET image reconstruction
Static PET images were reconstructed from 10-minute emission data (50-60 minutes after injection) using Siemens’ E7tools with ordered subset expectation maximization (OSEM), which was used as reference full-count PET images. Low-count PET data (10% count) were generated through Poisson thinning of the same listmode data.
Unsupervised learning image recovery
The network architecture used was a U-Net as shown in Figure 1. Similar to previous unsupervised learning framework1, anatomical MPRAGE images were used as input to the network and were trained to output low-count PET images. However, when stopped early before the network fully converges, the network will output a PET image similar to full-count images. As a result, the choice of stopping criteria is crucial.
In this work, we proposed an MRI-guided stopping criteria using the structural similarity index measure (SSIM) between the recovered PET images and anatomical MRI, and the mean absolute error (MAE) between the recovered PET images and the low-count PET images. The stopping criteria were also used as the loss function.
Evaluation
Results from the unsupervised learning U-Net framework were compared to full-count data to assess the best stopping criteria using the following quantitative metrics: 1) Peak Signal to Noise Ratio (PSNR), 2) SSIM, 3) Root Mean Square Error (RMSE), all with respect to the full-count image. One random subject was picked to tune the network including the hyperparameters and select the weight of SSIM and MAE in the loss function. The same hyperparameters and weighting were then used for all other data.
To compare with Gaussian denoising. The optimal Gaussian filter parameter was chosen using the same subject that was used to tune our model and the parameter was used on the rest of the dataset.
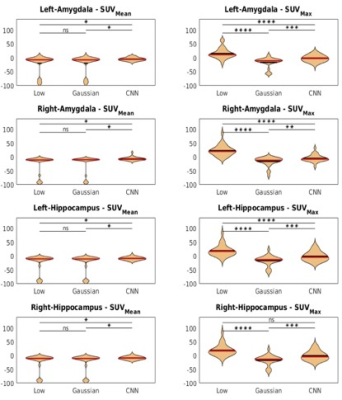
In addition to the quantitative image metrics, standard uptake value (SUV) was also calculated from low-count PET, network output, and Gaussian denoised PET. The mean SUV (SUVmean) and maximum SUV (SUVmax) were calculated for the hippocampus and amygdala (parcellation was performed using MPRAGE with Freesufer 7.0). The percentage SUV difference was computed using full-count PET as the reference. Paired t-test was performed to compare quantitative metrics and SUV differences.
Results
An example of network output is shown in Figure 2 from a representative subject.The PSNR, SSIM, and RMSE of the low-count PET, Gaussian denoised PET, and our unsupervised denoising network output for the subjects are shown in Figure 3. Paired student t-test demonstrated significant differences for all three metrics when comparing our network output to low-count and Gaussian denoised to network output. This demonstrates that our network output has improved global image similarity metrics.
The calculated SUVmean and SUVmax for the two ROIs are shown in Figure 4. As demonstrated in the figure, our network output also has improved SUV accuracy when compared to the full-count PET. Only the SUVmax of the right hippocampus showed no significance between low-count PET and CNN output. However, the CNN output still has a better standard deviation (SD) of 16.74 while low-count PET has a SD of 35.20.
Discussion
Our result shows that our novel stopping criteria for this unsupervised learning denoising network outperformed conventional gaussian filtered denoising and low-count PET in all global image similarity metrics with statistical significance.The SUV value also confirms this conclusion. We will further evaluate our novel stopping criterion together with our denoising network on other PET tracers utilizing simultaneous PET/MRI acquisition.
Acknowledgements
This work was supported by the National Institute of Mental Health (R01MH104512).References
1. Cui J, Gong K, Guo N, Wu C, Meng X, Kim K, et al. PET image denoising using unsupervised deep learning. Eur J Nucl Med Mol Imaging. 2019 Dec;46(13):2780–9.
2. Poynton CB, Chen KT, Chonde DB, Izquierdo-Garcia D, Gollub RL, Gerstner ER, et al. Probabilistic atlas-based segmentation of combined T1-weighted and DUTE MRI for calculation of head attenuation maps in integrated PET/MRI scanners. :12.
3. Chen KT, Izquierdo-Garcia D, Poynton CB, Chonde DB, Catana C. On the Accuracy and Reproducibility of a Novel Probabilistic Atlas-based Generation for Calculation of Head Attenuation Maps in Integrated PET/MR Scanners. Eur J Nucl Med Mol Imaging. 2017 Mar;44(3):398–407.
Figures
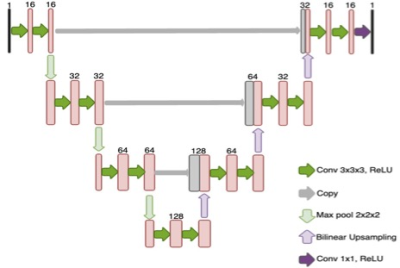
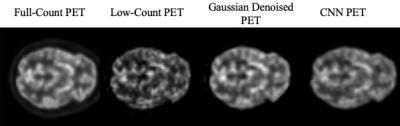
Figure 2. Example of low-count PET, Gaussian denoised PET, CNN recovered PET, and full-count PET.
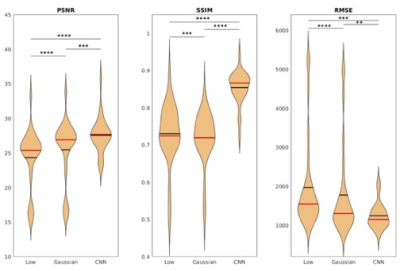
Figure 3. Violine plot of the PSNRa, SSIMa, and RMSEb for low-count PET, Gaussian denoised PET, and CNN output. ahigher is better, blower is better. *: P≤0.05, **:P≤0.01, ***:P≤0.001, ****:P≤0.0001, the black line indicates mean, the red line indicates median.