0872
A Time-Saving View-Sharing Interleaved Block-Segmented Diffusion-Tensor Imaging (VS iblocks-DTI)1Department of Biomedical Engineering, The Chinese University of Hong Kong, Shatin, Hong Kong, 2Multi-Scale Medical Robotics Center, Shatin, Hong Kong, 3Graduate Institute of Biomedical Electronics and Bioinformatics, National Taiwan University, Taipei, Taiwan, 4Department of Biomedical Engineering, University of Arizona, Tucson, AZ, United States, 5Brain Imaging and Analysis Center, Duke University Medical Center, Durham, NC, United States
Synopsis
Keywords: Image Reconstruction, Diffusion Tensor Imaging
Recently, a novel self-navigated multi-shot EPI data acquisition scheme called iblocks-EPI was proposed to achieve high-resolution DWI images with further reduction of distortion compared to the conventional interleaved EPI. However, the total acquisition time of iblocks-EPI is several times longer than the conventional interleaved EPI because it requires multiple patch patterns for covering a complete k space. Consequently, it is time-consuming and impractical for routine acquisition of DTI data. In this work we propose a view-sharing iblocks-DTI scheme which can substantially reduce the scan time for iblocks-DTI acquisition while providing accurate DTI tensor calculation.
Introduction
High-resolution multi-shot DTI is desirable for clinical applications and neuroscience research because it can improve the fiber delineation and brain network detections1,2. Recently, a self-navigated interleaved block-segmented EPI (iblocks-EPI)3 was demonstrated to be capable of generating high-resolution DWI images with several advantages: 1) the shortened echo-spacing (ESP) can further reduce geometric distortion compared to interleaved EPI; 2) the self-navigation of iblocks-EPI can make the inter-shot phase variation correction more robust for the data acquired at high magnetic fields or high b-values; 3) the oversampling of central k-space can improve the SNR performance of data reconstruction. However, iblocks-EPI requires five different patch patterns for covering a complete k-space, resulting in a substantial increase of scantime (i.e., 5-fold of conventional interleaved EPI). Therefore, using iblocks-EPI for DTI acquisition is relatively time-consuming and less practical, especially for collecting a large number of diffusion directions. Propeller imaging encounters similar challenges because it requires the acquisition of a significantly large number of blades. To improve the acquisition efficiency or temporal resolution, the view-sharing was combined with Propeller-EPI for Q-ball imaging or Propeller-GRE for dynamic-enhanced MRI in literature4,5. In this work, we combined view-sharing with iblocks-EPI to accelerate the DTI acquisition, thereby improving scan efficiency.Materials and Methods
View-sharing iblocks-DTI (VS iblocks-DTI) acquisition schemeIn previously proposed iblocks-EPI3, the whole 2D k-space is divided into 9 blocks (i.e., 3x3), and sampled by a total of 5 different patch patterns (patch pattern #1 ~ #5 in Fig.1). Each patch pattern consists of three blocks, including the central block for self-navigation. Owing to inconsistent off-resonance effects, central blocks in patch patterns #4 and #5 are only used for the navigation purpose. Fig.1 demonstrates the view-sharing acquisition scheme for the proposed VS iblocks-DTI. Patterns #1, #2, and #3 are acquired consecutively and repeatedly for varying diffusion-weighted directions since they include central blocks (colored in red, green, and blue in Fig.1) contributing to the image contrast, and these patterns are called target patterns. In addition, patch pattern #4 or #5 is acquired once every three diffusion directions to provide some necessary peripheral high-frequency data. Finally, for each diffusion direction, the central k-space block is filled only using the corresponding target pattern while other k-space blocks are filled from both the target pattern and its adjacent patterns acquired with different diffusion directions (Fig.1). Thus, at least three blocks of k-space data are acquired with the desired diffusion encoding for attaining the diffusion contrast and the other remaining blocks are acquired with different diffusion directions. To maintain the similarity of diffusion contrast among the shared k-space blocks, a smooth ordering of diffusion directions is adopted (Fig.2).
Experiments
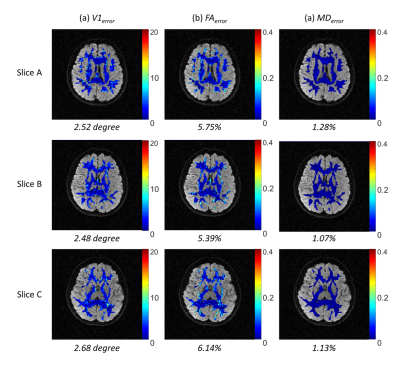
Hybrid simulations were performed to investigate the feasibility of the proposed VS iblocks-DTI. Firstly, a set of 4-shot interleaved DTI data with 64 diffusion directions was acquired on a 1.5 T MRI scanner using an 8-channel coil with following scan parameters: TE/TR = 72.4(full-echo)/4000 ms, FOV = 24×24 cm2, matrix size = 128×128, and b-value = 750 s/mm2. This 4-shot interleaved DTI data was reconstructed using MUSE6 and served as the gold standard of DTI tensor calculation. Secondly, a set of VS iblocks-DTI data was generated according to the patch patterns and acquisition scheme shown in Fig.1. Here an acquisition acceleration factor R_acq is defined as the ratio of the number of patterns need to be acquired in VS iblocks-DTI compared with fully-sampled iblocks-DTI, and R_acq equals to 0.268 (i.e., 73% reduction in scantime) for acquiring 64 diffusion directions using the view-sharing scheme shown in Fig.1. Afterward, the simulated VS iblocks-DTI data were regrouped accordingly and then reconstructed using the POCSMUSE3,7 for producing DWI image at each diffusion direction. Finally, the calculated DTI tensor was quantitatively evaluated with three error indexes by being compared to gold standard: 1) percentage fractional anisotropy (FA) deviation FAerror, 2) percentage mean diffusivity (MD) deviation MDerror, and 3) the angular deviation of the principal eigenvectors V1error.
Results and Discussion
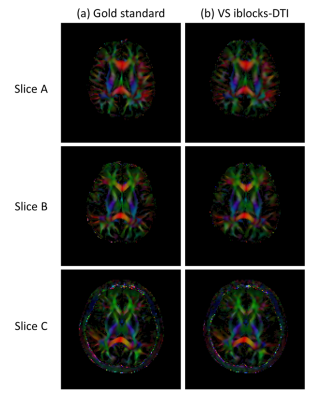
Fig.3 shows the comparison of six representative DWI images between gold standard and VS iblocks-DTI. Compared with gold standard, VS iblocks-DTI can produce DWI images with comparable quality, similar diffusion contrast, and small reconstruction errors. Fig.4 demonstrates that the DTI tensor produced by VS iblocks-DTI has a high similarity to the gold standard. Fig.5 shows that the averaged quantitative tensor calculation errors are minor for all V1, FA, and MD error maps (corresponding to 2.48~2.68°, 5.39~6.14%, and 1.07~1.28%, respectively).All the minor deviations between the VS iblocks-DTI results and the gold standard suggest that the proposed view-sharing scheme can substantially improve the acquisition efficiency for iblocks-DTI, without sacrificing image quality or accuracy in tensor calculation. In addition, VS iblocks-DTI can still retain the further reduced geometric distortion advantage of iblocks-EPI technique for producing high-quality DTI data. Furthermore, the view-sharing scheme proposed in this work can dramatically reduce the scan time of fully-sampled iblocks-DTI by 73%, which in turn requires only 1.34 times the scan time of conventional interleaved EPI (the fully-sampled iblocks-DTI: 5 times). In summary, the proposed VS iblocks-DTI can improve the feasibility of high-quality multi-shot DTI data with reasonable scantime, thereby providing an alternative for the robust multi-shot DTI acquisition strategy.
Acknowledgements
The work was in part supported by grants from Hong Kong Research Grant Council (GRF17106820, GRF17125321, and ECS24213522).References
1. Song AW, Chang H-C, Petty C, Guidon A, Chen N-K. Improved Delineation of Short Cortical Association Fibers and Gray/White Matter Boundary Using Whole-Brain Three-Dimensional Diffusion Tensor Imaging at Submillimeter Spatial Resolution. Brain Connect. 2014;4(9):636-640. doi:10.1089/brain.2014.02702. Chang HC, Sundman M, Petit L, et al. Human brain diffusion tensor imaging at submillimeter isotropic resolution on a 3Tesla clinical MRI scanner. Neuroimage. 2015;118:667-675. doi:10.1016/j.neuroimage.2015.06.016
3. Chang H-C, Chu M-L, Sundman M, Chen N-K. High-Quality and Self-Navigated Diffusion-Weighted Imaging Enabled by a Novel Interleaved Block-Segmented (iblocks) EPI. In: International Society for Magnetic Resonance in Medicine, 23rd Annual Meeting, Toronto, Ontario, Canada.
4. Chou M, Huang T, Chung H, Hsieh T, Chang H, Chen C. Q‐ball imaging with PROPELLER EPI acquisition. NMR Biomed. 2013;26(12):1723-1732.
5. Chuang T, Huang H, Chang H, Wu M. View‐sharing PROPELLER with pixel‐based optimal blade selection: Application on dynamic contrast‐enhanced imaging. Med Phys. 2014;41(6Part1):62302.
6. Chen N kuei, Guidon A, Chang HC, Song AW. A robust multi-shot scan strategy for high-resolution diffusion weighted MRI enabled by multiplexed sensitivity-encoding (MUSE). Neuroimage. 2013;72:41-47. doi:10.1016/j.neuroimage.2013.01.038
7. Chu M-L, Chang H-C, Chung H-W, Truong T-K, Bashir MR, Chen N. POCS-based reconstruction of multiplexed sensitivity encoded MRI (POCSMUSE): A general algorithm for reducing motion-related artifacts. Magn Reson Med. 2015;74(5):1336-1348. doi:10.1002/mrm.25527
Figures
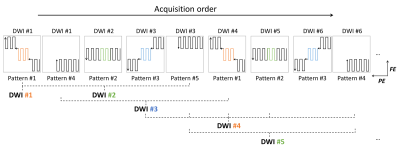
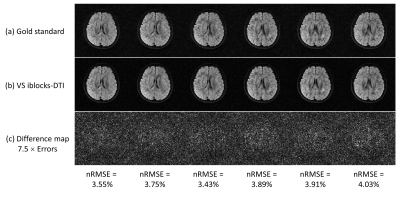
Fig. 3 Six representative DWI images from (a) MUSE-reconstructed gold standard, (b) POCSMUSE-reconstructed VS iblocks-DTI, and (c) their corresponding difference maps. The normalized root mean square errors (nRMSE) were calculated using: nRMSE = (√(1/N×∑((IVS-Ig)^2)))/(max(Ig)-min(Ig)), where Ig and IVS represent pixel intensities of DWI images from the gold standard and VS iblocks-DTI, respectively. N represents the number of all pixels.