0837
Investigating VoxelMorph Image Registration for Fast 4D Respiratory Compensated Image Reconstruction on an MR-Linac1Institute of Cancer Research, London, United Kingdom, 2Imperial College London, London, United Kingdom
Synopsis
Keywords: Image Reconstruction, Radiotherapy
Respiratory resolved (4D)-MRI is expected to benefit MRI-guided radiotherapy for abdominal-thoracic cancers. However, a current limitation of 4D-MRI is that one has to trade-off between artefacts, spatial-temporal resolution, and spatial coverage. Deformable image registration has been employed to enhance image quality of undersampled 4D-MRIs, but such approaches are usually too slow to be used in MR-guided radiotherapy workflows. We investigated the feasibility of employing models trained using the VoxelMorph framework with a view of leveraging a fast computation to minimise 4D-MRI reconstruction time on the MR-Linac. The models performed well and were ~48 times faster than a common registration method.Introduction
MRI-guided radiotherapy for abdominal-thoracic cancers is expected to benefit from respiratory-resolved (4D)-MRI.1 However, one current limitation of 4D-MRI is that one has to trade off between artefacts, spatial-temporal resolution, and spatial coverage.2 While higher spatial resolution allows for better definition of the tumour extent, sufficient temporal resolution is required to characterise the motion. Sufficient spatial coverage is required for treatment re-optimisation and to simultaneously monitor the positions of tumour and nearby radio-sensitive organs, such as the heart.Deformable image registration has been employed previously3 to enhance image quality of undersampled 4D-MRI data, but such approaches can take several hours to reconstruct due to many deformable registrations. Since reconstruction times for MR-guided radiotherapy workflows must be on the order of minutes4 (while the patient waits on the treatment couch), machine-learning-based approaches are likely to become key components of 4D-MRI for guided Radiotherapy.5
In this work, we investigate the feasibility of using image registration models trained using the VoxelMorph framework6 with a view of leveraging a fast computation to minimise reconstruction time for MR-guided radiotherapy. Because the respiratory-compensated (MoCo) image reconstruction developed by Rank et al.3 uses image registrations at multiple resolution levels, we train networks for a range of resolution levels.
Methods
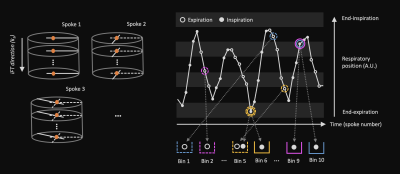
Thirty-four free-breathing, golden-angle, radial 3D-VANE images were acquired on a 1.5-T MR-Linac system (Unity, Elekta AB, Stockholm) with two 4-channel phased array coils for 11 patient volunteers (4.8 min per scan). A large field-of-view (500x500x200 mm) was centred on the liver, with a 1.61 oversampling factor along z, 332 spokes, and 75 kz partitions. A range (1-6) of image volumes was acquired for each patient, where an even split of the datasets was acquired without and without radiotherapy compression belts.Respiratory resolved, 4D-MRI images were reconstructed offline in MATLAB (Mathworks, Natick, MA) at 3 isotropic resolution levels: 40x40x16 (~12 mm), 80x80x32 (~6 mm), and 160x160x64 (~3 mm). Figure 1 illustrates our approach to respiratory binning: A self-navigation signal was derived from the inverse Fourier transform along kz of the central point of each k-space spoke using principal component analysis. Following classification of expiration and inspiration time points, data were sorted by respiratory signal into 10 respiratory bins with a 50% data overlap of adjacent bins. High-quality images were reconstructed using the non-uniform Fourier transform and compressed sensing (XD-GRASP7).
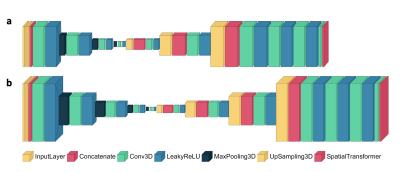
These respiratory-resolved images were used to train models with the VoxelMorph framework to learn 3D deformation vector fields (DFVs). Three models were trained, corresponding to each resolution level. For each network, the default framework hyper-parameters were used (learning rate=0.0001, regularisation rate=0.01) with a mean-squared-error-based loss. The shapes of the networks are shown in Figure 2. For training, a batch size of 8 was used and 150 epochs and 50 validation steps per epoch were chosen. In total, the VoxelMorph framework was trained with 24 4D volumes (22,680 image pairs, 9 patients), validated with 5 4D volumes, (4725 image pairs, 1 patient), and tested with 5 4D volumes (1 patient). The models were evaluated by comparison to a Demons-like algorithm8 for image registration in MATLAB, where this category of image-registration technique was used previously for MoCo image reconstruction.3
Results and Discussion
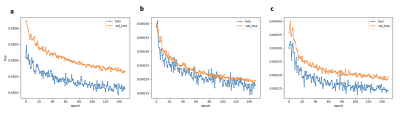
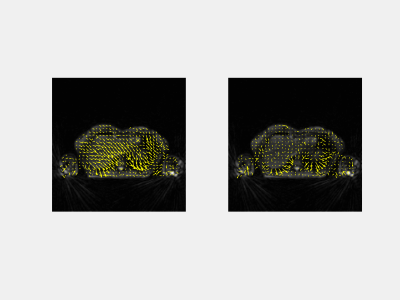
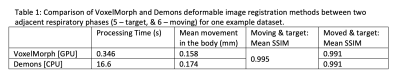
The training times for each network were: ~0.75 h, ~1.5 h, and ~6 h, respectively. Figure 3 illustrates the loss over epochs for each model. Figure 4 illustrates the x-y components of the 3D DFVs generated for a test patient at the level of the diaphragm. Comparing DVFs generated from the Demons-like algorithm with those from VoxelMorph visually, distinctions are apparent although broadly similar patterns, with comparable vector magnitudes, are also seen. The magnitude of motion averaged voxelwise over the body is ~0.2 mm for both methods (Table 1). In this test case, both registration techniques also produced SSIMs between the moved and fixed image volumes of 0.991.The processing time for GPU VoxelMorph image registration between adjacent respiratory phases was found to be ~48 times faster (0.3 s for a 160x160x64 volume) than for a commonly used CPU Demons registration algorithm. This rapid registration time is similar to durations achieved in work by Terpstra et al.9 on machine-learning-based image registration for undersampled thoracic MR images. In our apprach, very few changes were made to the default parameters of the VoxelMorph framework.
Conclusion and Future Work
In conclusion, the VoxelMorph framework could be a convenient and useful tool for developing cutting-edge 4D-MRI techniques for MRI-guided radiotherapy. In the next stage of work, we will further refine and validate the image registration training and use the resulting models to build an offline reconstruction method similar to the method implemented by Rank et al.:3 a faster MoCo 4D-MRI approach. Long image registration times in MoCo were a major bottleneck of total reconstruction times. By transferring the computational burden to offline training, we hope to develop an optimal 4D-MRI approach for the MR-Linac.Acknowledgements
The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are members of the Elekta MR-Linac Research Consortium. We acknowledge research support from Elekta and Philips MR for the source codes and required research licenses. We recieve funding from Cancer Research UK programme grant C33589/A28284.References
1. Godley A, Zheng D and Rong Y. MR-linac is the best modality for lung SBRT. J Appl Clin Med Phys. 2019;20:7-11.
2. Freedman J, Bainbridge H, Nill S, et al. Synthetic 4D-CT of the thorax for treatment plan adaptation on MR-guided radiotherapy systems. Phys Med Biol. 2019;64:115005.
3. Rank C Thorsten H, Buzan M, et al. 4D respiratory motion-compensated image reconstruction of free-breathing radial MR data with very high undersampling. Magn Reson Med. 2016;77:1170-1183.
4. Mickevicius N and Paulson E. Investigation of undersampling and reconstruction algorithm dependence on respiratory correlated 4D-MRI for online MR-guided radiation therapy. Phys Med Biol. 2017;62:2910.
5. Stemkens B, Paulson E and Tijssen R. Nuts and bolts of 4D-MRI for radiotherapy. Phys Med Biol. 2018;63:21TR01.
6. Balakrishnan G, Zhao A, Sabuncu M, et al. VoxelMorph: A Learning Framework for Deformable Medical Image Registration. IEEE Trans Med Imaging, 2019;38.
7. Feng L, Axel L, Chandarana H, et al. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med. 2016;75 (2):775-88.
8. Vercauteren T, Pennec X, Perchant A, et al. Diffeomorphic demons: efficient non-parametric image registration. NeuroImage 2009;45:S61–S72.
9. Terpstra M, Maspero M, Bruijnen T, et al. Real‐time 3D motion estimation from undersampled MRI using multi‐resolution neural networks. Med Phys. 2021;48(11):6597–6613.
Figures