0836
Attention-Based Multi-Offset Deep Learning Reconstruction for Accelerating Chemical Exchange Saturation Transfer MRI1Department of Biomedical Engineering, City University of Hong Kong, Hong Kong, Hong Kong, 2Hong Kong Centre for Cerebro-Cardiovascular Health Engineering (COCHE), Hong Kong, Hong Kong, 3City University of Hong Kong Shenzhen Research Institute, Shenzhen, China, 4Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 5Tung Biomedical Sciences Centre, City University of Hong Kong, Hong Kong, Hong Kong
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence
We proposed an attention-based multi-offset network to exploit redundant anatomy information for the reconstruction of CEST-MR image (AMO-CEST). To the best of our knowledge, this is the first work using deep learning with varied radial sample patterns and multi-offset slices as input to accelerate CEST-MRI. Compared with other deep learning-based methods on the four times under-sampling mouse brain CEST dataset, the AMO-CEST achieved the best performance with an MMSE of , a PSNR of dB, and an SSIM . In conclusion, the proposed AMO-CEST network can accelerate the CEST-MRI at high down-sampling rate while maintaining good image quality.Introduction
Chemical exchange saturation transfer (CEST) magnetic resonance imaging (MRI) is a promising imaging technique that can non-invasively detect molecular information and thereby has been applied in imaging many diseases, such as cancer, stroke, Alzheimer's disease, and multiple sclerosis 1-7. Relatively long scan time is one of the major challenges of applying CEST-MRI in clinics. This is because CEST-MRI image at multiple saturation offsets are required for extraction of molecular information. To accelerate the CEST-MRI, several reconstruction methods have been proposed 8-12. However, the similarity of anatomical structural features at different frequency offsets has not been utilized in these studies. Based on this, we proposed to use the varied and complementary radial masks to under-sample the k-space at multi-offset. To make use of the multi-offset input, we developed the attention-based multi-offset network (AMO-CEST) to reconstruct high-quality CEST slices from down-sampling slices.Method
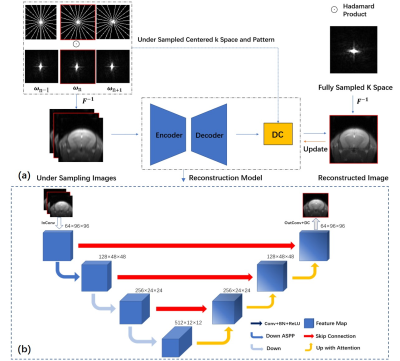
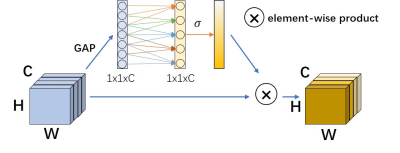
Five C57BL/6 male mice were used in this study. MRI was performed on a 3T Bruker BioSpec system. CEST datasets were acquired using a continuous-wave saturation module followed by a RARE readout. For each mouse, CEST data under 2 saturation powers (0.8 and 1.2 mT) from 3 orientations (2 slices for each) were acquired. Each CEST dataset includes 76 slices with 73 CEST slices (-20 to 20 ppm) and 3 M0 slices (200 ppm). Hence, the total number of CEST images and slices were 60 and 5760, respectively. The structure of AMO-CEST is shown in Figure 1. Varied and complementary radial masks with the least intersection are designed for CEST images at different adjacent offsets. Specifically, the adjacent radial mask rotates at a specific degree to meet the requirement. This could help the neural network model acquire more information between offsets, as shown in Figure 1(a). ωn-1,ωn and ωn+1 represent a group of three adjacent offsets. The under-sampled MRI k-space data is computed by Hadamard product the fully sampled k space with the designed radial masks. As illustrated in Figure 1 (b), the structure of AMO-CEST contained is an encoder-decoder structure, which has three downsampling and upsampling operations, with skip connection to enlarge the spatial receptive field. The AMO-Net mainly consists of three parts: atrous spatial pyramid pooling (ASPP) 13, channel attention module 14,15, and data consistency (DC) module 16. Both real and imaginary parts of MR slices are used for reconstruction. Hence, the network input size is 2C×Nx×Ny, while the output size is 2×Nx×Ny, where C=3 is the number of adjacent offsets. Figure 2 illustrates the channel attention module which could explore the multi-offset information and assign different weights to the different feature map channels. The data consistency module could preserve the original information and avoid degrading prediction results.Results and Discussion
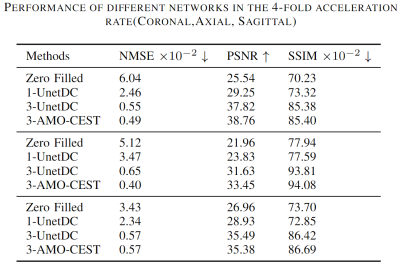
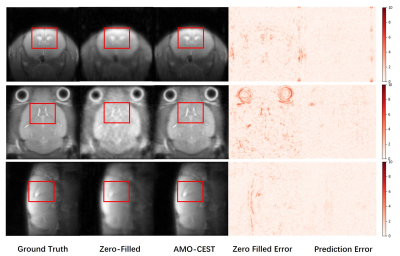
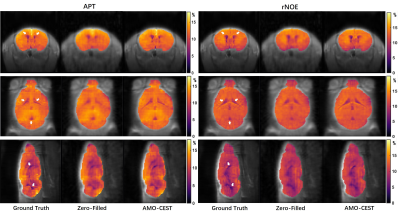
The 60 CEST datasets were split into 51 and 9 for training and testing, respectively. We used normalized mean squared error (NMSE), peak signal-to-noise ratio (PSNR), and structural similarity index measurement (SSIM) to assess the network performance. Table 1 summarized the results of zero-filling and all reconstruction models at 4-fold down-sampling. Except for the zero-filling, the method name is described as #-model name, where "#" is referring to the number of adjacent slices used as input. Three rows from top to bottom were the results of coronal, axial, and sagittal orientations, respectively. Compared to the zero-filling results, all deep learning methods could effectively improve the reconstructed image quality for three orientations even though data of different orientations have different metric scores at the same under-sampling rate. Furthermore, by comparing the 1-UnetDC and 3-UnetDC, we observed that the strategy of using three adjacent slices at multi-offset as inputs could effectively improve image quality. Notably, the results of the three orientations support that our proposed AMO-CEST model achieved the best performance among these methods. For example, in coronal orientation, the proposed AMO-CEST showed the best performance with an NMSE of 0.49 , a PSNR of 35.86 and an SSIM of 88.72x10-2, which are much higher than those of zero-filling operation and other models. Figure 3 and 4 indicate the visual improvement in raw CEST slices and extracted CEST maps, respectively, by using AMO-CEST reconstruction. For CEST maps, amide proton transfer (APT) at 3.5 ppm and relayed nuclear Overhauser effect (rNOE) at -3.5 ppm were extracted for comparison. Brain structures, such as cerebrospinal fluid (boxes in Figure 3 and arrows in Figure 4), are blurred in zero-filling slice but well reconstructed in AMO-CEST predicted slice. Both quantitative and visual results demonstrate our proposed AMO-CEST could effectively reconstruct high-quality slice from down-sampling slice.Conclusion
We proposed an attention-based multi-offset network (AMO-CEST) with a radial-based multi-offset sample strategy. This AMO-CEST utilizes the redundant structure information of slices at multi-offset. The quantitative and visual results indicate that our proposed method could achieve superior results compared to existing methods. As far as we know, this is the first work using deep learning with varied radial sample patterns and multi-offset slices as input to reconstruct CEST image. Moreover, this sampling technique is practical and effective in CEST acquisition. AMO-CEST has the potential to accelerate CEST-MRI and facilitate its clinical translation.Acknowledgements
Authors would like to acknowledge the funding supports from Research Grants Council (11102218, PDFS2122-1S01, 11200422, RFS2223-1S02, C1134-20G); City University of Hong Kong (7005433, 7005626, 9239070, 9609307, 9610560); National Natural Science Foundation China (81871409); Tung Biomedical Sciences Centre; Hong Kong Centre for Cerebro-cardiovascular Health Engineering. This work was carried out using the computational facilities, CityU Burgundy, managed and provided by the Computing Services Centre at City University of Hong Kong (https://www.cityu.edu.hk/)References
1. Van Zijl, Peter CM, and Nirbhay N. Yadav. "Chemical exchange saturation transfer (CEST): what is in a name and what isn't?." Magnetic resonance in medicine 65.4 (2011): 927-948.
2. Zhou, Jinyuan, et al. "Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI." Nature medicine 9.8 (2003): 1085-1090.
3. Zhou, Jinyuan, et al. "Amide proton transfer (APT) contrast for imaging of brain tumors." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 50.6 (2003): 1120-1126.
4. Huang, Jianpan, et al. "Molecular Imaging of Brain Tumors and Drug Delivery Using CEST MRI: Promises and Challenges." Pharmaceutics 14.2 (2022): 451.
5. Sun, Phillip Zhe, et al. "Relaxation‐compensated fast multislice amide proton transfer (APT) imaging of acute ischemic stroke." Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 59.5 (2008): 1175-1182.
6. Huang, Jianpan, et al. "Altered d-glucose in brain parenchyma and cerebrospinal fluid of early Alzheimer’s disease detected by dynamic glucose-enhanced MRI." Science advances 6.20 (2020): eaba3884.
7. Huang, Jianpan, et al. "Relayed nuclear Overhauser effect weighted (rNOEw) imaging identifies multiple sclerosis." NeuroImage: Clinical 32 (2021): 102867.
8. Doneva, Mariya, et al. "Compressed sensing reconstruction for magnetic resonance parameter mapping." Magnetic Resonance in Medicine 64.4 (2010): 1114-1120.
9. Wang, Shanshan, et al. "Accelerating magnetic resonance imaging via deep learning." 2016 IEEE 13th international symposium on biomedical imaging (ISBI). IEEE, 2016.
10. Xu, Jianping, et al. "Accelerating Chemical Exchange Saturation Transfer Imaging Using a Model-based Deep Neural Network." arXiv preprint arXiv:2205.10265 (2022).
11. Guo, Chenlu, et al. "Fast chemical exchange saturation transfer imaging based on PROPELLER acquisition and deep neural network reconstruction." Magnetic Resonance in Medicine 84.6 (2020): 3192-3205.
12. Wang, Puyang, et al. "Improving amide proton transfer-weighted mri reconstruction using t2-weighted images." International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2020.
13. He, Kaiming, et al. "Spatial pyramid pooling in deep convolutional networks for visual recognition." IEEE transactions on pattern analysis and machine intelligence 37.9 (2015): 1904-1916.
14. Hu, Jie, Li Shen, and Gang Sun. "Squeeze-and-excitation networks." Proceedings of the IEEE conference on computer vision and pattern recognition. 2018.
15. Wang, Qilong, et al. "Supplementary material for ‘ECA-Net: Efficient channel attention for deep convolutional neural networks." Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition, IEEE, Seattle, WA, USA. 2020.
16. Schlemper, Jo, et al. "A deep cascade of convolutional neural networks for MR image reconstruction." International conference on information processing in medical imaging. Springer, Cham, 2017.
Figures