0835
DeepSTI: Towards Tensor Reconstruction using Fewer Orientations in Susceptibility Tensor Imaging1The Johns Hopkins University, Baltimore, MD, United States
Synopsis
Keywords: Image Reconstruction, Susceptibility, Susceptibility Tensor Imaging
The application of STI in human in vivo has been practically infeasible because of its time-consuming acquisition scheme. We propose a novel image reconstruction algorithm for STI that leverages data-driven priors to tackle this issue. Our method, called DeepSTI, learns the data prior implicitly via a deep neural network that resembles the proximal operator of a regularizer function. The dipole inversion problem is then solved iteratively using the learned proximal network. Experimental results demonstrate superior performance of DeepSTI over state-of-the-art methods. DeepSTI is the first reconstruction method to achieve high quality results for human STI with fewer than six orientations.Introduction
Susceptibility tensor imaging (STI) is an emerging magnetic resonance imaging technique that characterizes the anisotropic tissue magnetic susceptibility in vivo with high spatial resolution [1]. It has the potential to provide information for the reconstruction of white matter fiber pathways as well as detection of myelin changes in the brain, which is of great value for understanding brain function under normal and pathological conditions. However, the application of STI has been hindered by its cumbersome and time-consuming acquisition protocol, which requires scans at multiple head orientations (usually >6) for sufficient information to solve the ill-posed dipole inversion problem. Furthermore, the limited head rotation angles due to physical constraints of the head coil pose a significant challenge for the inversion problem even when a large number of orientations have been acquired. As a result, STI has not yet been widely applied in humans in vivo. In this work, we aim to tackle these issues by proposing a novel image reconstruction algorithm for STI that leverages data-driven priors, allowing for susceptibility tensor estimation from fewer orientations and closing the gap to deploying STI in vivo in clinical settings.Methods
Our method, called DeepSTI, learns a data prior implicitly via a deep neural network that approximates the proximal operator of the regularizer function for STI. The dipole inversion problem is then solved iteratively using the learned proximal network with fewer head orientations. We formulate the STI dipole inversion problem as a regularized linear inverse problem$\min_{\x} \frac{1}{2} \|\M(\y - \F^{-1}\mathcal{A}\F\x)\|_2^2 + R(\x),$
where x∈R^6n is the underlying tensor image (symmetric 2nd order tensor, n is the number of voxels), y∈R^mn denotes the observed local field change (m is the number of head orientations), F denotes discrete Fourier Transform, A is the dipole kernel, and M is a brain mask. Further, f(x)=1⁄2 ‖M(y-F^(-1) AFx)‖_2^2:R^6n→R is a data fidelity term. The regularizer, R(x):R^6n→R, is a suitable function that introduces prior knowledge and allows for a robust reconstruction. We follow a proximal gradient approach by taking the gradient of the differentiable function f(x) and the proximal of the regularizer, defined as P_R. In our method, to leverage the advantage of deep neural networks for learning powerful priors from large amounts of data, we parameterize the proximal operator P_R by a deep neural network G_θ, which produces the learned proximal iterates given by:
$x_{k+1} & = G_\theta(x_k - \alpha_k \nabla f(x_k))$
where α_k∈R is the step size at the k-th iteration and θ denotes all trainable parameters of the neural network. Further details of implementation can be found in Ref [8].
To train the network, we leveraged a supervised learning framework by minimizing the distance between estimated and ground-truth STI over all training samples. To increase the diversity of training data and further improve the performance, data augmentation is performed, including random affine (scaling and rotation) and elastic transform, adding random-sized cuboid-shape isotropic lesions at random locations, and training with a random number of orientations (between 1 and 6).
For in vivo data acquisition, GRE measurements were acquired from 8 human subjects, including 3 subjects on a 3T scanner with 1.5 mm isotropic resolution as in Ref. [2] and 5 subjects on a 7T scanner with 1 mm isotropic resolution, reconstructed to 0.98×0.98×1 mm3 spatial resolution as in Ref. [3]. We also obtained DTI data (b-value of 700 s/mm2, 32 diffusion encoding, NSA=2) from all the subjects at 2.2 mm isotropic resolution, reconstructed and interpolated to similar resolution as the GRE scans. The study was approved by the local IRB and all volunteers signed informed consent.
Results and Discussion
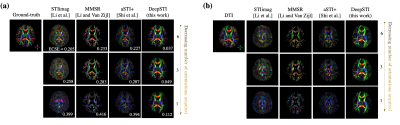
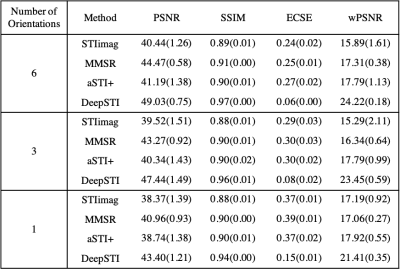
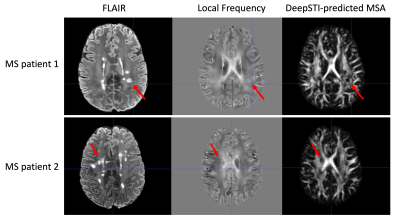
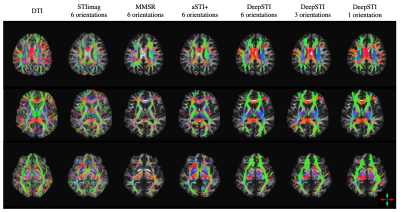
We evaluated DeepSTI on both simulation and in vivo data and compared it with state-of-the-art methods including STIimag [4], MMSR [2] and aSTI+ [5]. Fig. 1 shows the principal eigenvector (PEV) maps estimated from simulation (a) and in vivo measurements (b) acquired from a testing subject at 3T and 1.5 mm isotropic resolution. A significant improvement can be observed in the results yielded by DeepSTI. Note that with only a few (e.g., 1∼2) head orientations, DeepSTI can still yield a reasonable estimation of the PEV map, while the other methods fail to provide any reasonable estimation. Figure 2 summarizes the quantitative metrics for STI reconstruction from 2 testing subjects, where our method consistently outperforms other methods in various metrics. Figure 3 depicts MSA maps calculated from DeepSTI using only one head orientation for two multiple sclerosis (MS) patients, showing a decrease in MSA in lesion regions. Figure 4 depicts STI-based fiber tracking results using in vivo data from a 3T subject. It is clear from these that our method offers more coherent and complete tracking results than other alternatives, demonstrating the potential of STI-based fiber tractography with DeepSTI.Conclusion
We proposed DeepSTI, a new method for STI dipole inversion that enables STI reconstructions with measurements at fewer head orientations by leveraging data-driven priors. Experimental results from simulation and in vivo human brain data demonstrate the superiority of DeepSTI compared to state-of-the-art methods for susceptibility tensor reconstruction, fiber direction estimation and fiber tractography. Our results shed light on the potential of large-scale application of STI on human in vivo for better understanding brain functions and neurological diseases.Acknowledgements
This research has been supported by NIH Grant P41EB031771, as well as by the Toffler Charitable Trust and by the Distinguished Graduate Student Fellows program of the KAVLI Neuroscience Discovery Institute.References
[1] Liu C. Susceptibility tensor imaging. Magn. Reson.Med. 2010;63(3):1471-1477.
[2] Li, X. and Van Zijl, P.C., 2014. Mean magnetic susceptibility regularized susceptibility tensor imaging (MMSR-STI) for estimating orientations of white matter fibers in human brain. Magnetic Resonance in Medicine, 72(3), pp.610-619.
[3] Li, X., et al, 2012. Mapping magnetic susceptibility anisotropies of white matter in vivo in the human brain at 7T. NeuroImage 62, 314-330.
[4] Li, W., et al, 2017. Susceptibility tensor imaging (STI) of the brain. NMR in Biomedicine, 30(4), p.e3540.
[5] Shi, Y., Cao, S., Li, X., Feng, R., Zhuang, J., Zhang, Y., Liu, C. and Wei, H., 2022. Regularized Asymmetric Susceptibility Tensor Imaging in the Human Brain in Vivo. IEEE Journal of Biomedical and Health Informatics, 26(9), pp.4508-4518.
[6] Lai, K.W., et al, 2020. Learned proximal networks for quantitative susceptibility mapping. In MICCAI (pp. 125-135).
[7] Lee, K., et al, 2017. Superhuman accuracy on the SNEMI3D connectomics challenge. arXiv preprint arXiv:1706.00120.
[8] Fang, Z., Lai, K.W., van Zijl, P., Li, X. and Sulam, J., 2022. DeepSTI: Towards Tensor Reconstruction using Fewer Orientations in Susceptibility Tensor Imaging. arXiv preprint arXiv:2209.04504.
Figures