0834
NPB-REC: Non-parametric Assessment of Uncertainty in Deep-learning-based MRI Reconstruction from Undersampled Data1The Interdisciplinary Applied mathematics Program, Technion, Haifa, Israel, 2Faculty of Biomedical Engineering, Technion, Haifa, Israel
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence
Deep learning (DL) models are currently employed to reconstruct high quality MRI image from undersampled k-space measurements. Yet, uncertainty quantification in images reconstructed by such models is critical for reliable clinical decision making. We propose NPBREC, a non-parametric Bayesian approach for uncertainty estimation in DL-based MRI reconstruction. We demonstrated the added-value of our Bayesian registration framework on the fastmri multi-coil brain MRI dataset, compared to the baseline E2E-VarNet trained with and without inference-time dropout for uncertainty quantification. Our NPBREC approach demonstrated both improved reconstruction accuracy and a better correlation between reconstruction errors and uncertainty measures.Introduction
Reducing MRI acquisition time by under-sampling the ``k-space'' (i.e. Fourier domain) constitutes a key necessity in enabling advanced MRI applications such as cardiac and fetal imaging. Further, acceleration of MRI will also reduce MRI vulnerability to patient motion during the acquisition process. However, the under-sampled data results in aliasing artefacts in the reconstructed images. Recently, deep learning (DL) based models are successfully used for MRI reconstruction from undersampled data [2,3]. However, their practical utilization in neuroimaging or other clinical applications is questionable as they lack of computational mechanisms for quantifying the risks of failures. It is therefore essential to develop uncertainty aware reconstruction approaches, which will enable a safer utilization of DL models in clinical applications. Motivated by this fact, we tackle the reconstruction task from a Bayesian perspective [4] and propose NPBREC, a non-parametric Bayesian approach for uncertainty assessment in MRI reconstruction from undersampled k-space data. The proposed approach can provide quantitative measures of uncertainty correlated with risk of failure in the MRI image predictions, and can improve the overall generalization of the image reconstruction.Methods
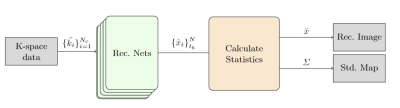
The backbone of our reconstruction system is based on the E2E-VarNet model [2]. We use Stochastic gradient Langevin dynamics (SGLD) [5] during the training phase to enable uncertainty estimation by characterizing the posterior distribution of the network weights. Specifically, we used noise injection for the training loss gradients to efficiently sample the true posterior distribution.Fig.1 illustrates the operation of the NPB-REC system at the inference phase. Firstly, we sample a set of reconstructed images obtained by feed-forwarding the under-sampled k-space data to the reconstruction models with the weights obtained by training with SGLD in the last 9 epochs. Then, when we have a new image to reconstruct, we estimate the averaged posterior image. Further, we quantify the std. of the reconstruction, which is used to characterize the pixel-wise uncertainty map. We demonstrated the added-value of our approach on the multi-coil MRI dataset, taken from the fastmri challenge [1]. In our experiments, we train our model on the multi coil brain data. At inference, we use annotated images of both multi coil Brain and Knee, which were taken from fastmri+ [7]. The Knee database is used to assess the generalization ability of our model in anatomical shift. The training and validation sets include 4469 and 1113 images of size 16x 320x 320. For inference, we use 247 and 155 annotated images from the brain and knee datasets, respectively. From these datasets, we generate 71504, 17808 and 5792 2D images for the training, validation and inference phases, respectively
Results and Discussion
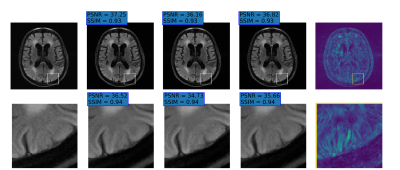
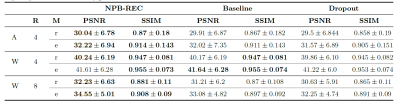
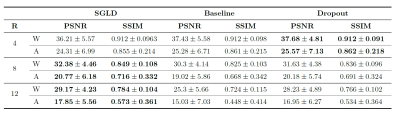
Fig.2 presents examples of reconstruction results obtained by our NPB-REC approach, the baseline, and Monte Carlo Dropout. We assessed the performance of our model by means of PSNR and SSIM, in comparison to the baseline E2E-VarNet [2] with and without inference-time dropout [6]. As depicted in Table 1, our experiments show that NPB-REC outperforms the baseline by means of reconstruction accuracy (PSNR and SSIM of 34.55, 0.908 vs. 33.08, 0.897, p<0.01) in high acceleration rates (R=8). This is also measured in regions of clinical annotations. The improvement in the reconstruction performance can be noted both quantitatively from the metrics especially for masks with acceleration rate R=8 and qualitatively via the images of annotations, where our results shows less smoothness than that obtained by Dropout. Further, our approach enables better generalization of the reconstruction performance in high acceleration rates for knee dataset, even when the model is trained only on brain data (Table 2, R=8,12). We calculated the mean value of the Std. maps, obtained by our method and Monte Carlo Dropout, for all images in the inference set and utilize it as uncertainty measure. The correlation between these uncertainty measures and reconstruction error (MSE) are depicted in Fig.3. Our method provides a more accurate estimate of the uncertainty that correlates with the reconstruction error, compared to the Monte-Carlo inference time Dropout method [3] (Pearson correlation coefficient of R=0.94 vs. R=0.91). These outcomes, in turn, indicate the ability of our uncertainty measure to detect unreliable reconstruction performance.Conclusions
We developed NPB-REC, a non-parametric Bayesian method for the reconstruction of brain MRI images from under-sampled k-space data with uncertainty estimation. The conducted experiments showed that our approach enables uncertainty quantification that exhibits higher correlation with the reconstruction error than that obtained by Monte Carlo Dropout. Our non-parametric Bayesian approach has the potential to facilitate safe utilization of DL based methods for MRI reconstruction from undersampled data. It provides principled mechanisms to quantify the risks of failures in the DL-based model predictions, which is necessary in the safety-critical applications. The proposed NPB-REC improves generalization both in high acceleration rates and anatomical shift, allows the assessment of the uncertainty in the predictions and provide a principled mechanism to determine out-of-distribution data.Acknowledgements
Khawaled, S. is a fellow of the Ariane de Rothschild Women Doctoral Program.References
[1] https://fastmri.org/
[2] Sriram A, Zbontar J, Murrell T, Defazio A, Zitnick CL, Yakubova N, Knoll F, Johnson P. End-to-end variational networks for accelerated MRI reconstruction. InInternational Conference on Medical Image Computing and Computer-Assisted Intervention 2020 Oct 4 (pp. 64-73). Springer, Cham.
[3] Shaul, Roy, et al. "Subsampled Brain MRI Reconstruction by Generative Adversarial Neural Networks." Medical Image Analysis (2020): 101747.
[4] Neal, Radford M. Bayesian learning for neural networks. Vol. 118. Springer Science & Business Media, 2012.
[5] Welling, Max, and Yee W. Teh. "Bayesian learning via stochastic gradient Langevin dynamics." Proceedings of the 28th international conference on machine learning (ICML-11). 2011.
[6] Y. Gal and Z. Ghahramani, “Dropout as a Bayesian approximation: Representing model uncertainty in deep learning,” in 33rd International Conference on Machine Learning, ICML 2016, 2016, vol. 3, pp. 1651–1660
[7 ] Zhao, Ruiyang, et al. "fastmri+: Clinical pathology annotations for knee and brain fully sampled multi-coil mri data." arXiv preprint arXiv:2109.03812 (2021).
Figures