0825
Deep-learning-based transformation of magnitude images to synthetic raw data for deep-learning-based image reconstruction1Department of Radiology and Nuclear Medicine, St. Olav's University Hospital, Trondheim, Norway, 2Department of Circulation and Medical Imaging, NTNU - Norwegian University of Science and Technology, Trondheim, Norway
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, synthetic data, compressed sensing, accelerated imaging
This study demonstrates using generative deep learning for transforming magnitude-only images into synthetic raw data for deep-learning-base image reconstruction. Using relatively few raw datasets, a set of neural networks was trained to generate the missing phase and coil sensitivity information in magnitude-only images. These maps are then recombined into synthetic raw data. We trained end-to-end variational networks for 4-fold accelerated compressed sensing reconstruction on the FastMRI dataset, with increasing training set size. Synthetic raw data showed similar improvements as real raw data with increasing data. This shows promise for applying deep-learning-based image reconstruction when raw data is scarce.Purpose
Deep-learning-based MR image reconstruction has shown great promise for accelerated imaging1,2. However, these systems are limited by the availability of raw data, which limits new applications and clinical adoption.The fastMRI dataset3,4 released 8400 raw MRI datasets, which enables training of complex reconstruction models with “big” data. However, in both research and clinical practice, raw data is rarely stored, which makes gathering these amounts of raw datasets for new applications virtually impossible. Reconstructed, magnitude-only images are more easily accessible, both in research datasets and in hospital databases. However, using magnitude-only data for training deep-learning-based reconstruction can lead to mismatches between the training data and actually acquired raw data, and thus poor generalization5. Synthetically generating the missing phase and coil sensitivity information is often based on simplified models, which fail to capture the complexity of these maps.
Here, we investigate using deep learning to generate synthetic raw data from magnitude-only images, to alleviate the raw data requirements of deep-learning-based image reconstruction, building on our previous work6. We trained neural networks to generate raw data using a limited number of raw datasets from the fastMRI database, and transformed larger magnitude-only datasets into synthetic raw data, which was used to train deep-learning-based compressed sensing reconstructions.
Methods
DataWe selected 184 (160 train, 4 validation, 20 test) T1-weighted FLASH brain scans from the NYU fastMRI database4, acquired at 3 tesla (flip angle=70, TE=2.64, TR=250, TI=300, matrix=16x320x320). These scans were acquired using 16 (head) or 20 (head+neck) receive coils, of which we maintained the 16 head coils, and calculated coil sensitivity maps (CSMs)7.
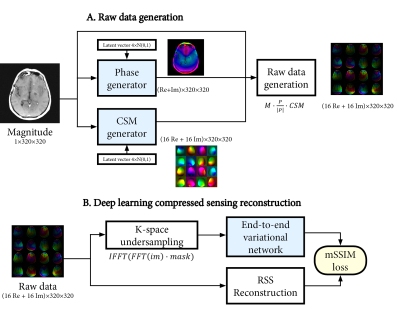
Raw data generation
We trained a set of neural networks to generate realistic phase and CSM maps from a magnitude-only image. Combined with the magnitude-only image, synthetic raw data can then be generated as:
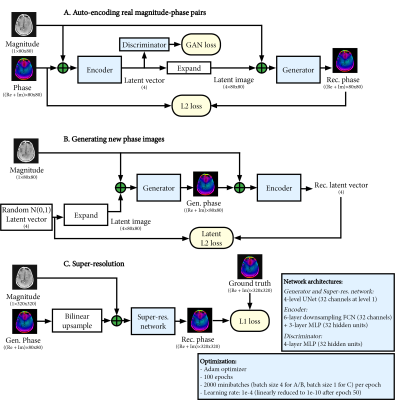
$$raw_{gen}=M\cdot~P_{gen}\cdot~CSM_{gen}$$ The generative networks were based on a conditional adversarial auto-encoder architecture8. This first stage (Figure 1A/B) generates low-resolution phase and CSM maps (4-fold downsampled, 80x80). The auto-encoder learns the variability in the phase and CSM, and can generate new samples through random sampling of a latent vector. In the second stage (Figure 1C), a UNet network9 upsamples the generated 80x80 phase and CSM maps back to the original 320x320 resolution. The networks in both stages were trained separately for phase and CSM generation, using only 20 raw datasets for training.
Image reconstruction
We demonstrate the value of synthetic raw data generation in a 4-fold multi-coil compressed sensing reconstruction experiment, identical to the FastMRI challenge (4). The reconstruction method was an end-to-end variational network (VarNet)10. Reconstructions were evaluated on the test set using the mean absolute error (MAE), root mean squared error (RMSE), and mean structural similarity index (mSSIM) metrics.
As a baseline, the VarNet was trained with 20, 50, 100, and 160 raw datasets. Using synthetic data generation, we then trained the VarNet with 20 raw datasets and 20, 50, 100, and 160 magnitude-only datasets. Synthetic raw data was randomly generated during training from the magnitude-only data, and noise was added according to the noise covariance observed in the raw data. Each experiment was repeated 5 times, and the repetition with the best validation performance was chosen for evaluation.
Results
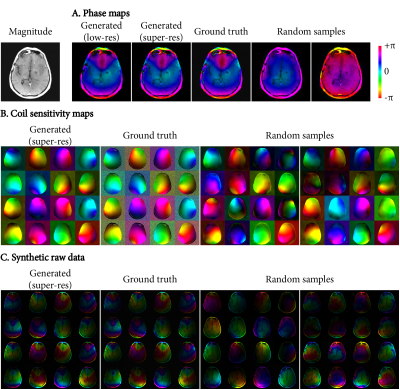
Figure 3 shows synthetic phase, CSM and raw data. Our method can both approximate the ground truth accurately, and simultaneously sample the variance present in the training dataset.Figure 4 shows the mean reconstruction errors on the test set for increasing data availability (real raw data for the baseline, magnitude-only images for the synthetic data experiment). For small data availability (20-50 scans), using synthetic data improved the reconstruction metrics by providing randomization in the synthetic raw data. For larger datasets (100+ scans), real raw data showed better scaling with increasing data, although synthetic data also continued to reduce errors at least up to 160 scans.
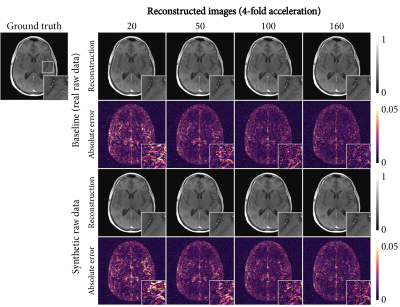
Reconstructed images are shown in Figure 5, which demonstrates the reduction of errors with increasing data availability for both the baseline and using synthetic raw data. Finer details were slightly better preserved for real raw data than for synthetic raw data.
Discussion
This study demonstrates that variability in raw datasets of limited size can be learned with deep learning and used to generate randomized, realistic phase and coil sensitivity maps at the full image resolution. Together with magnitude-only images, these maps recombine into synthetic raw data, which showed improved reconstruction errors as more magnitude-only images were available. Although using additional real raw data showed slightly better image quality, the cost and effort required to acquire raw data makes synthetic raw data based on magnitude-only images an appealing alternative.Synthetic raw data provides an MR-physics-inspired alternative to conventional data augmentation in deep learning. As data augmentation introduces potentially unrealistic image transformations, synthetic data may be preferred, though this needs to be investigated further. Furthermore, the two approaches could be combined.
This study shows that generative deep learning can improve deep-learning-based image reconstruction by transforming magnitude-only images into additional synthetic raw training data. Magnitude images should be more readily available and thus alleviate the raw data requirements. This is particularly promising for applying deep-learning-based reconstruction to new imaging sequences, where raw data is scarce.
Acknowledgements
This work was supported by the Research Council of Norway (FRIPRO Researcher Project 302624).References
1. Knoll F, Murrell T, Sriram A, et al. Advancing machine learning for MR image reconstruction with an open competition: Overview of the 2019 fastMRI challenge. Magn. Reson. Med. 2020;84:3054–3070
2. Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature 2018;555:487–492
3. Zbontar J, Knoll F, Sriram A, et al. fastMRI: An Open Dataset and Benchmarks for Accelerated MRI. arXiv:1811.08839, 2019.
4. Knoll F, Zbontar J, Sriram A, et al. fastMRI: A Publicly Available Raw k-Space and DICOM Dataset of Knee Images for Accelerated MR Image Reconstruction Using Machine Learning. Radiol. Artif. Intell. 2020;2:e190007
5. Shimron E, Tamir JI, Wang K, Lustig M. Subtle Inverse Crimes: Naïvely training machine learning algorithms could lead to overly-optimistic results. arXiv:2109.08237, 2021
6. Zijlstra F, While P. Deep-learning-based raw data generation for deep-learning-based image reconstruction. In: Proceedings of the 31st Annual Meeting of the ISMRM, no. 0049.; 2022.
7. Inati SJ, Hansen MS, Kellman P. A Fast Optimal Method for Coil Sensitivity Estimation and Adaptive Coil Combination for Complex Images. In: Proceedings of the 22nd Annual Meeting of the ISMRM.; 2014.
8. Makhzani A, Shlens J, Jaitly N, Goodfellow I, Frey B. Adversarial Autoencoders. arXiv:1511.05644, 2016
9. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science. Springer, Cham; 2015. pp. 234–241
10. Sriram A, Zbontar J, Murrell T, et al. End-to-End Variational Networks for Accelerated MRI Reconstruction. arXiv:2004.06688, 2020
Figures