0823
MERLIN: In-depth investigation on complex-valued image reconstruction in PyTorch and Tensorflow1Department of Radiotherapy, University Medical Center Utrecht, Utrecht, Netherlands, 2Computational Imaging Group for MR Diagnostics & Therapy, University Medical Center Utrecht, Utrecht, Netherlands, 3Lab for AI in Medcine, Technical University of Munich, Munich, Germany, 4Department of Computing, Imperial College London, London, United Kingdom, 5Medical Image And Data Analysis (MIDAS.lab), University Hospital of Tübingen, Tübingen, Germany
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence
Machine learning (ML) has become a powerful technique for reconstructing undersampled MRI. However, applying ML to MRI reconstruction requires several essential building blocks, consisting of general operations and MRI-specific operators in the context of reconstruction. While the former are available in generic frameworks such as Keras/TensorFlow or PyTorch, the MR-specific operators are generally custom-implemented. MERLIN was proposed as a generic toolkit for ML-based medical imaging to harmonize the machine learning landscape and to provide complex-valued interfaces for commonly used back-ends. Here, we evaluate MERLIN as a cross-platform toolkit for complex-valued image reconstruction.Introduction
Machine learning (ML) has become a powerful technique for reconstructing undersampled MRI. However, applying ML to MRI reconstruction requires several essential building blocks, consisting of general operations (e.g., data loading and preprocessing, (complex-valued) operators, optimization, and backpropagation) and MRI-specific operators in the context of reconstruction (e.g., Fourier transformations, coil sensitivity maps, and data consistency). While the former are available in generic frameworks such as Keras/TensorFlow or PyTorch, the MR-specific operators are generally custom-implemented, as the complex-valued nature of MRI also demands handling this information. Previous works have addressed this as either complex-valued operations or as 2-channel real-valued operations[1]. MERLIN[2] was proposed as a generic toolkit for ML-based medical imaging to harmonize the ML landscape and to provide complex-valued interfaces for commonly used back-ends. Even though the processing mode and choice of framework can impact the model performance, thorough studies have not yet been performed. In this work, we evaluate MERLIN for ML platform-independent MRI reconstruction and research the impact of activation functions, loss functions, ML framework, and using complex-valued operations on reconstructed image quality.Methods
Four components of deep neural networks were evaluated: complex-valued versus real-valued operations, the activation function, the loss function, and the deep learning framework.Model architecture & training
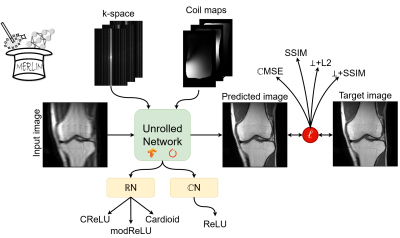
Complex-valued and two-channel real-valued unrolled networks[3] (ℂN and ℝN, respectively) with data consistency were implemented using the MERLIN framework in Tensorflow and PyTorch (Figure 1). These networks consisted of 10 cascades with four convolutional layers per cascade. The ℂN used 16, 16, 8, and 1 filters per layer, respectively, while the ℝN used 16, 16, 8, and 2 filters per layer, respectively. For ℂN, we evaluated the Cardioid[4], ℂReLU[5], and modReLU[6] activation functions, while for the ℝN, we used the ReLU activation function. We evaluated the MSE, DSSIM (i.e., 1-SSIM), ⊥+L2[7], and ⊥+DSSIM as loss functions. All models were trained for ten epochs using the AdamW optimizer (lr=1e-3, weight decay=1e-4) on a subset of the fastMRI dataset[8] (i.e., only the images without fat-suppression and 368 phase encode lines were used) with a batch size of three using an acceleration factor of 4 (Magic mask[9], ACS region fraction = 0.04). The k-space and target images were normalized such that the target image has a magnitude between 0 and 1. Coil-sensitivity maps were computed using ESPIRiT[10], and complex Gaussian noise (N(0,0.025)) was added to the undersampled k-space. Finally, models implemented in PyTorch and Tensorflow were overfitted on a single subject to establish the maximum attainable performance without any sources of random variations (i.e., disabling data shuffling, identical weight initialization, discarding random noise, deterministic training).
Evaluation
Performance for all models was evaluated using the magnitude SSIM, magnitude PNSR, and the phase RMSE (PRMSE) on the fastMRI validation set. Statistical significance was established using the Wilcoxon signed-rank test ($$$\alpha<0.01$$$).
Results
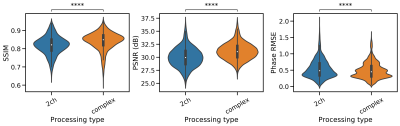
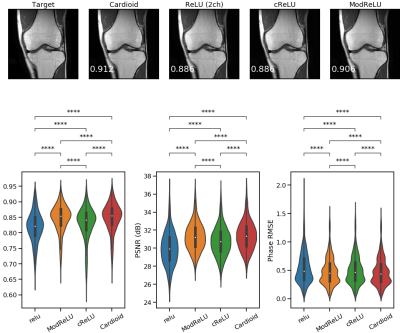
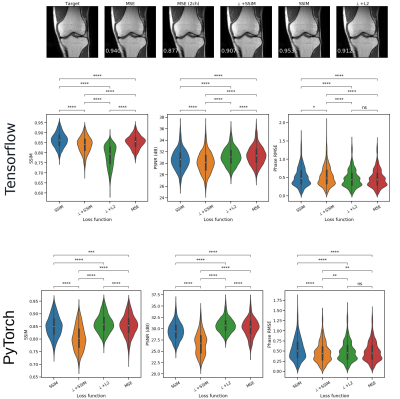
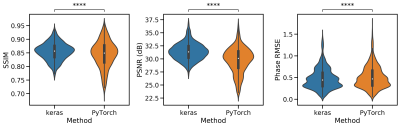
In all runs, we observed that the ℂN outperformed the ℝN (Fig. 2, SSIM 0.84±0.05 vs 0.81±0.05, Wilcoxon p < 0.01). The best-performing activation function was the Cardioid, closely followed by the modReLU loss function (Fig. 3). Based on our results, we observed that the performance of the MSE as a loss function was on par with the SSIM loss function (0.86±0.03 vs. 0.83±0.04). However, in PyTorch, ⊥+L2 performed better than all other loss functions, while the MSE was the best-performing loss function when using Tensorflow (Fig. 4). In general, we observed that Tensorflow demonstrated higher performance than PyTorch (Fig. 5, SSIM 0.834±0.03 vs 0.828±0.050, Wilcoxon p < 0.01). When overfitting on single subjects, Tensorflow outperforms PyTorch in controlled experiments (SSIM 0.929±0.03 vs. 0.919±0.02).Discussion
We have evaluated MERLIN for ML-based image reconstruction. By evaluating ℂN and ℝN on publicly-available data, we have found that complex-valued ML models outperform real-valued ML models, which aligns with previous results[11]. Moreover, we identified that the Cardioid and modReLU non-linear activation functions perform best for these models, which aligns with previous literature[4]. However, these operations have only been applied to models operating in the image domain. Future work might investigate whether an image-only, k-space, or hybrid ML model is the best approach. However, we noticed a discrepancy between the performance of ⊥-Loss in Tensorflow and PyTorch. In PyTorch, this loss function performs better than the MSE, which is not the case in Tensorflow. Moreover, we have independently verified that for this problem, with this dataset and this model architecture, all models implemented in Tensorflow outperform those same models implemented in PyTorch. Given the widespread use of these libraries, this is cause for concern. However, we have yet to identify the root cause of this issue and cannot conclude where this discrepancy arises. However, this observation identifies the need for a unified MRI-ML framework, as results should depend on methodology rather than on the choice of framework. As far as we know, no work has compared implementations between frameworks, possibly precluding observation of this effect. We call upon the community to investigate whether similar issues are observed in other applications and plan to benchmark this further.Conclusion
We have investigated MERLIN, a cross-platform MRI-ML library, for complex-valued image reconstruction. Here, we found that complex-valued ML models outperform real-valued ML models. Moreover, we identified a discrepancy in the performance between Tensorflow and PyTorch.Acknowledgements
This work is part of the SIGNET project, which is a project in the ITEA program. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Quadro RTX 5000 GPU used for prototyping this research. We would like to thank all current and future MERLIN contributors.References
[1] Hammernik et al. "Physics-Driven Deep Learning for Computational Magnetic Resonance Imaging." arXiv preprint arXiv:2203.12215 (2022).
[2] Hammernik and Küstner. Proc. Int. Soc. Mag. Res. Med. (2022)
[3] Hammernik et al. "Learning a variational network for reconstruction of accelerated MRI data." Magnetic resonance in medicine 79.6 (2018): 3055-3071.
[4] Virtue et al. "Better than real: Complex-valued neural nets for MRI fingerprinting." Proc. IEEE international conference on image processing (ICIP) (2017) (pp. 3953-3957).
[5] Scardapane et al. "Complex-valued neural networks with nonparametric activation functions." IEEE Transactions on Emerging Topics in Computational Intelligence 4.2 (2018): 140-150.
[6] Arjovsky et al. "Unitary evolution recurrent neural networks." International conference on machine learning. PMLR (2016).
[7] Terpstra et al. "⊥-loss: a symmetric loss function for magnetic resonance imaging reconstruction and image registration with deep learning." Medical Image Analysis (2022): 102509.
[8] Zbontar et al. "fastMRI: An open dataset and benchmarks for accelerated MRI." arXiv preprint arXiv:1811.08839 (2018).
[9] Defazio, Aaron. "Offset sampling improves deep learning based accelerated mri reconstructions by exploiting symmetry." arXiv preprint arXiv:1912.01101 (2019).
[10] Uecker et al. "ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA." Magnetic resonance in medicine 71.3 (2014): 990-1001.
[11] Cole et al. "Analysis of deep complex‐valued convolutional neural networks for MRI reconstruction and phase‐focused applications." Magnetic resonance in medicine 86.2 (2021): 1093-1109.
Figures