0819
Attention mechanisms for sharing low-rank, image and k-space information during MR image reconstruction1Medical Image and Data Analysis (MIDAS.lab), Department of Diagnostic and Interventional Radiology, University Hospital of Tuebingen, Tuebingen, Germany, 2Lab for AI in Medcine, Technical University of Munich, Munich, Germany, 3Department of Computing, Imperial College London, London, United Kingdom, 4Max Planck Institute for Intelligent Systems, Tuebingen, Germany
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, Image Reconstruction, Heart
Cardiac CINE MR imaging requires long acquisitions under multiple breath-holds. With the development of deep learning-based reconstruction methods, the acceleration rate and reconstructed image quality have been increased. However, existing methods face several shortcomings, such as limited information-sharing across domains and generalizability which may restrict their clinical adoption. To address these issues, we propose A-LIKNet which incorporates attention mechanisms and maximizes information sharing between low-rank, image, and k-space in an interleaved architecture. Results indicate that the proposed A-LIKNet outperforms other methods for up to 24x accelerated acquisitions within a single breath-hold.Introduction
Cardiac CINE MR imaging allows for accurate and reproducible measurement of cardiac function. Conventionally, multi-slice 2D CINE images are acquired under multiple breath-holds leading to patient discomfort and imprecise assessment. With the development of deep learning-based reconstruction methods, the acceleration rate and reconstructed image quality have been increased.However, we observe following challenges for learning-based methods:
1. Operation on single domain: image enhancement1-3, low-rank4,5, and k-space learning6,7 focus on reconstruction either in the image or k-space. Although physics-based unrolled networks8-11 leverage k-space information in the data consistency (DC) layer, k-space information is not learned specifically.
2. Focus on single-domain regularization: most image-based networks1-3,8-11 ignore the inherent low-rank property of dynamic imaging12 which can be leveraged4,5.
3. Equal feature contribution: features along coils or time are treated equally, impairing the representation ability. Attention mechanisms were studied13-15, but focused on spatial or network-channel-wise attention.
These challenges may limit the full potential of deep learning physics-based reconstruction. Thus we hypothesize that the reachable acceleration rate and reconstructed image quality still have room for improvement.
In this work, we incorporate attention mechanisms in a physics-based unrolled reconstruction network that leverages low-rank, image and k-space information, named A-LIKNet. We maximize the information sharing between domains by parallel k-space and image branches in an interleaved architecture with learnable information-sharing layer (ISL). As dynamic CINE images have a strong low-rank property, both sparse and low-rank priors are learned. Furthermore, attention along time is applied in the image domain to improve dynamic delineation, and attention along coils learns to weigh the coils in k-space domain. Experiments show that the proposed A-LIKNet can effectively reconstruct up to 24x accelerated cardiac CINE images, enabling single breath-hold imaging.
Methods
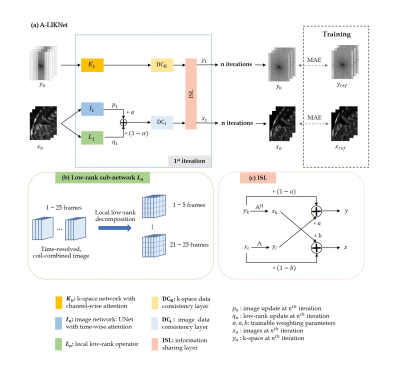
The overall structure of the proposed network is depicted in Fig.1(a), consisting of a k-space branch, an image branch, and ISL.The network is trained to solve the following problem: $$\mathbf{x}=\arg\,\underset{\mathbf{x}}{\min}\ \frac{1}{2}\parallel\mathbf{Ax}-\mathbf{y}\parallel_{2}^{2}+\lambda_{1}R_{1}\left(\mathbf{x}\right)+\lambda_{2}R_{2}\left(\mathbf{x}\right)$$ which incorporates a sparse $$$R_{1}()$$$ and a low-rank $$$R_{2}()$$$ regularization with weightings $$$\lambda_{1}$$$ and $$$\lambda_{2}$$$ to reconstruct the image $$$\mathbf{x}$$$ from the acquired k-space $$$\mathbf{y}$$$. The encoding operator $$$\mathbf{A}$$$ is composed of the Fourier transformation, undersampling trajectory and coil sensitivities.
The problem is reformulated using variable splitting with auxiliary variables $$$\mathbf{p}$$$ and $$$\mathbf{q}$$$ into the unconstrained Lagrangian function: $$\textit{L}_{\mu_{1},\mu_{2}}(\mathbf{x},\mathbf{p},\mathbf{q})=\frac{1}{2}\parallel\mathbf{Ax}-\mathbf{y}\parallel_{2}^{2}+\lambda_{1}R_{1}\left(\mathbf{p}\right)+\lambda_{2}R_{2}\left(\mathbf{q}\right)+\frac{\mu_{1}}{2}\left\|\mathbf{p}-\mathbf{x}\right\|^{2}_2+\frac{\mu_{2}}{2}\left\|\mathbf{q}-\mathbf{x}\right\|^{2}_2$$ which can be solved iteratively: $$\left\{\begin{matrix}\mathbf{p}_{n+1}={\arg}\,\underset{\mathbf{p}}{\min}\,\frac{\mu_{1}}{2}\left\|\mathbf{p}-\mathbf{x}_{n}\right\|^{2}_2+\lambda_{1}R_{1}\left(\mathbf{p}\right)\\\mathbf{q}_{n+1}={\arg}\,\underset{\mathbf{q}}{\min}\,\frac{\mu_{2}}{2}\left\|\mathbf{q}-\mathbf{x}_{n}\right\|^{2}_2+\lambda_{2}R_{2}\left(\mathbf{q}\right)\\\mathbf{x}_{n+1}={\arg}\,\underset{\mathbf{x}}{\min}\,\frac{1}{2}\left||\mathbf{Ax}-\mathbf{y}\right\|_{2}^{2}+\frac{\mu_{1}}{2}\left\|\mathbf{p}_{n+1}-\mathbf{x}\right\|^{2}+\frac{\mu_{2}}{2}\left\|\mathbf{q}_{n+1}-\mathbf{x}\right\|^{2}\end{matrix}\right.$$ with the sub-problems being formulated as learnable regularizers: $$\left\{\begin{matrix}\mathbf{p}_{n+1}=\textbf{I}_{n+1}(\mathbf{x}_{n})\\\mathbf{q}_{n+1}=\textbf{L}_{n+1}(\mathbf{x}_{n})\\\mathbf{x}_{n+1}=\textbf{DC}_\text{I}(\alpha\cdot\mathbf{p}_{n+1}+(1-\alpha)\cdot\mathbf{q}_{n+1},\mathbf{A},\mathbf{y})\end{matrix}\right.$$ for an image sub-network $$$\textbf{I}_n$$$, low-rank sub-network $$$\textbf{L}_n$$$ weighted by a trainable parameter $$$\alpha=\frac{\mu_{1}}{\mu_{1}+\mu_{2}}$$$ (initialized at 0.5) in a data consistency layer $$$\textbf{DC}_\text{I}$$$ (gradient descent) in image domain.
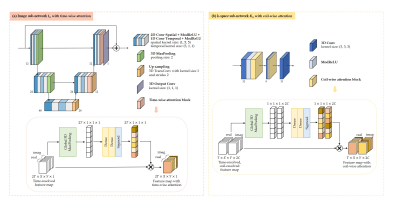
The image sub-network $$$\textbf{I}_{n}$$$ (Fig.2(a)) is expressed as a complex-valued residual 2D+t UNet with attention blocks along time in the decoder. Specifically, time-resolved and spatial-channel-squeezed information is extracted and encoded by one 3D pooling and two fully-connected layers to generate the attention map which will excite the decoder features.
The low-rank sub-network $$$\textbf{L}_{n}$$$ (Fig.1(b)) learns singular value thresholds $$$\lambda_{i}$$$ for local spatial-temporal patches of 4x4 neighbourhood in 5 frames, following13: $$\textbf{L}(\mathbf{x}_{n})=\mathbf{U}\cdot\operatorname{diag}\left(\max\left(\operatorname{sigmoid}\left(\lambda_{i}\right)\ast\max\left(\sigma_{in}\right),\sigma_{in}\right)\right)\cdot\mathbf{V}^{H}$$ with singular values of $$$i^{th}$$$ patch $$$\sigma_{in}\,(1\le n\le \operatorname{rank}\left(\mathbf{x}_{i}\right)$$$) to update the whole time sequence $$$\mathbf{q}$$$ which is merged with the image update $$$\mathbf{p}$$$ by $$$\alpha$$$. $$$\textbf{DC}_\text{I}$$$ performs gradient descent data consistency on this combined output to update the image branch.
The k-space sub-network (Fig.2(b)) uses attention along coils in a three-layer architecture with 3D complex-valued convolutions along spatial-temporal dimensions and a null-space projection in k-space $$$\mathbf{DC}_\text{K}$$$.
While local information is captured by the image branch, feature information on a global scale and across coils is captured in the k-space branch. Outputs of the k-space and image branch (after data consistency) are combined in an information sharing layer (Fig.1(c)). Data cross-over between domains is encoded via the respective forward and adjoint encoding operators and weighted by a trainable parameter.
Investigations are carried out on 2D cardiac CINE (bSSFP, TE/TR=1.06/2.12ms, $$$\alpha$$$=52°, resolution=1.9x1.9mm2, slice thickness=8mm) acquired on a 1.5T MRI in 129 subjects (38 healthy subjects and 91 patients). Data were split into 115 training and 14 unique test subjects. VISTA16 undersampling with random acceleration factors 2x to 24x is used. The proposed network was trained with Adam (learning rate=$$$10^{-4}$$$, batch size=1). The proposed method is compared to compressed sensing ($$$l_{1}$$$-wavelet regularization)17 and MoDL9. Furthermore, the effect of the k-space branch is investigated in the A-LINet, i.e. representing the network without k-space branch.
Results and Discussion
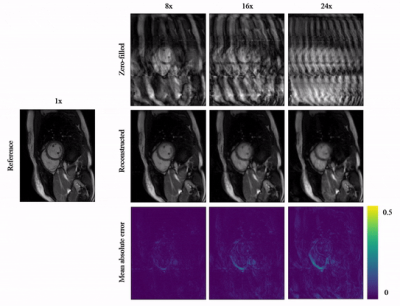
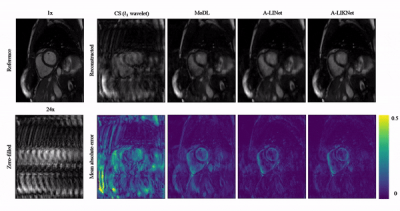
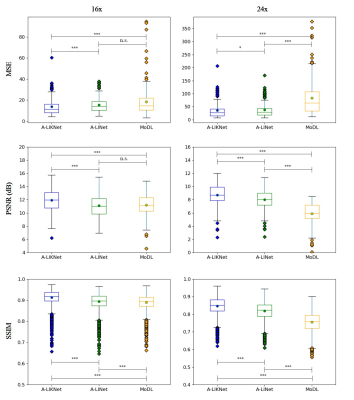
Animated Fig.3 shows reconstructions with the proposed A-LIKNet for 8x, 16x and 24x undersampled 2D cardiac CINE of one patient with active myocarditis, respectively. We can observe stable and effective reconstruction under different accelerations.A comparison to other methods and the ablated A-LINet is shown in Fig.4 for 24x acceleration. CS can hardly reconstruct images under this high acceleration rate. A-LIKNet shows less error than MoDL, and presents sharper details with improved contrast compared to A-LINet. Fig.5 evaluates the quantitative assessment (MSE, PSNR, SSIM) for MoDL, A-LINet, and A-LIKNet over all test subjects under 16x and 24x acceleration. The proposed A-LIKNet outperforms other networks regarding all metrics.
Conclusion
The proposed A-LIKNet with attention mechanism and ISL maximizes the information sharing between both image/k-space and spatial-temporal domains. As a result, A-LIKNet can efficiently reconstruct 2D cardiac CINE under high acceleration that would enable single breath-hold imaging.Acknowledgements
S.G. and T.K. contributed equally.References
1. D. Lee et al., "Deep Residual Learning for Accelerated MRI using Magnitude and Phase Networks", IEEE Trans on Biomedical Engineering 2018; 65(9):1985-95.
2. A. Hauptmann et al., "Real-time cardiovascular MR with spatio-temporal artifact suppression using deep learning–proof of concept in congenital heart disease", Magn Reson Med 2019; 81:1143-56.
3. A. Kofler et al., "Spatio-Temporal Deep Learning-Based Undersampling Artefact Reduction for 2D Radial Cine MRI With Limited Training Data", IEEE Trans Med Imaging 2020; 39(3):703-17.
4. C. Sandino et al., "DSLR+: Enhancing deep subspace learning reconstruction for high-dimensional MRI", Proc Intl Soc Mag Reson Med 2021; 29.
5. W. Huang et al., "Deep low-Rank plus sparse network for dynamic MR imaging", Med Image Anal 2021; 73:102190.
6. D. Lee et al., "Acceleration of MR parameter mapping using annihilating filter-based low rank hankel matrix (ALOHA)", Magn Reson Med 2016; 76(6):1848-64.
7. M. Akçakaya et al., "Scan-specific robust artificial-neural-networks for k-space interpolation (RAKI) reconstruction: Database-free deep learning for fast imaging", Magn Reson Med 2019; 81:439-53.
8. J. Schlemper et al., "A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction", IEEE Trans Med Imaging 2018; 37(2):491-503.
9. H. Aggarwal et al., "MoDL: Model-Based Deep Learning Architecture for Inverse Problems", IEEE Trans Med Imaging 2019; 38(2):394-405.
10. K. Hammernik et al., "Dynamic multicoil reconstruction using variational networks", Proc ISMRM 27th Annu Meeting Exhibit 2019; 4656.
11. T. Küstner et al., "CINENet: deep learning-based 3D cardiac CINE MRI reconstruction with multi-coil complex-valued 4D spatio-temporal convolutions", Sci Rep 2020; 10(1):13710.
12. R. Otazo et al., "Low-rank plus sparse matrix decomposition for accelerated dynamic MRI with separation of background and dynamic components", Magn Reson Med 2015; 73(3):1125-36.
13. Q. Huang et al., "MRI reconstruction via cascaded channel-wise attention network", ISBI 2019; 1622-26.
14. G. Li et al., "A Modified Generative Adversarial Network Using Spatial and Channel-Wise Attention for CS-MRI Reconstruction", IEEE Access 2021; 9:83185-98.
15. G. Li et al., "High-Resolution Pelvic MRI Reconstruction Using a Generative Adversarial Network With Attentionand Cyclic Loss", IEEE Access 2021; 9:105951-64.
16. R. Ahmad et al., "Variable density incoherent spatiotemporal acquisition (VISTA) for highly accelerated cardiac MRI", Magn Reson Med 2015; 74(5):1266-78.
17. M. Lustig et al., "Sparse MRI: The application of compressed sensing for rapid MR imaging", Magn Reson Med 2007; 58(6):1182-95.
Figures