0817
Automatic Segmentation and Diagnosis of Breast Cancer in Multicenter Data based on Deep Learning and Tissue-specific Histogram Normalization1Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 2Philips Healthcare, Shanghai, China, 3Shanghai Cancer Center, Fudan University, Shanghai, China, 4Human Phenome Institute, Fudan University, Shanghai, China
Synopsis
Keywords: Segmentation, Breast
This study presented an intelligent diagnosis system to segment breast tumors in dynamic contrast-enhanced (DCE) images from multicenter dataset and determine the risk of triple-negative breast cancer (TNBC) with a four-step model: a) breast segmentation with no-new Unet (nnUnet); b) multicenter data normalization using Tissue-specific Histogram Normalization (TSHN); c) tumor segmentation with nnUnet model and d) automatic diagnosis of breast cancer with radiomics analysis based on segmented masks. The proposed model exhibited a superior performance in segmentation and diagnosis of breast cancer in multicenter data.
INTRODUCTION
Breast cancer is the second most common cancer diagnosed among women in the United States1, in which TNBC is characterized by high malignancy and poor prognosis. With the knowledge of exact subtype, doctors can design individualized treatment plans. However, the subtype of breast cancer is mainly determined by immunohistochemistry, which is invasive and time-consuming. Recently, computer-aided diagnosis show potential in the prediction of cancer subtypes2-4, but the generalization of computer-aided models may decrease when used in multicenter data. Our proposed intelligent system adopted deep learning and radiomics analysis to diagnose the risk of TNBC and we further used the TSHN algorithm to improve the diagnostic performance of the model in multicenter data.METHODS
This study included 844 cases from five sources: 612 cases from Center A (Aurora 1.5T), 99 cases from Center B, 133 cases from Center C with three scanners (GE 3.0T/Philips 3.0T/Siemens 3.0T). All data had manually labeled tumor masks but only data from Center B had pathological tumor subtype labels(38 patients with TNBC and 61 patients with non-TNBC). 428 cases from Center A were used to train deep learning network for tumor segmentation, and the rest of the multicenter data formed the test cohort. In addition, data from Center B were used to construct radiomics model for TNBC prediction and its performance was assessed by a five-fold cross-validation.The proposed model consisted of four steps (Figure 1): breast segmentation, multicenter data normalization, segmentation and diagnosis of breast cancer. In the segmentation part, we adopted nnUnet, a self-adapting deep learning framework based on U-net5, to segment breast and tumors. Specifically, the 3D full resolution U-Net mode was applied in nnUnet framework (Figure 2), and the segmented breast mask was used as the initial ROI for tumor segmentation. In the data normalization part, based on the segmented breast masks, TSHN algorithm was selected for eliminating the heterogeneity of imaging features of multicenter data, which aims to match the intensity of tissues without upsetting the natural balance of tissue intensities6. In the radiomics analysis part, we extracted radiomics features including shape, texture, wavelet … features using Pyradiomics package (https://github.com/Pyradiomics), and used the LightGBM algorithm for feature selection and classifier construction.
In terms of performance evaluation, the segmentation performance was evaluated using DICE coefficient. The diagnostic performance was assessed by quality metrics of accuracy, areas under the curve (AUC), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV).
RESULTS
Examples of breast and tumor segmentation are shown in Figure 3a and Figure 3b, respectively. Multicenter data normalization is gainful for improving tumor segmentation performance, an example comparison of qualitative segmentation results with and without the TSHN is shown in Figure 3c. Moreover, we compared TSHN with two other multicenter data normalization algorithms, including histogram equalization (HE), histogram specification (HS), and the results show that the TSHN algorithm has superior performance in improving the generalization performance of the segmentation model, with an increase in the DICE coefficient of 0.1558 for Center B and 0.0578 for Center C compared to the unnormalized model (Table 1).As for automatic diagnosis of TNBC, the proposed model achieved an AUC of 95.53% and accuracy of 88.04% for per-patient analysis in the test cohort. In addition, to verify the feasibility of automatic tumor segmentation and multicenter data normalization for subsequent diagnostic task, we compared the results of the radiomics analysis based on ROIs from manual segmentation (MS), proposed automatic segmentation with TSHN (PAS), and automatic segmentation without TSHN (AS). The results show that the proposed model with PAS achieves even better performance in the prediction of TNBC compared with manual segmentation, in which TSHN played a crucial role (Table 2).
DISCUSSION
The proposed model enables automated breast tumor segmentation and TNBC diagnosis with high accuracy, and the results show its feasibility in multicenter data processing. Localization of breast tumors is the initial step for tumor subtype diagnosis, however, most of the existing studies relies on manual or semi-automatic tumor annotation, which requires medical knowledge and is subjective7,8. Herein, we proposed an intelligent system to automatically segment breast tumors and diagnose the risk of TNBC. In order to improve the generalization of the model, a normalization process of the multicenter data is essential. We compared three different normalization methods, and the experiment proved TSHN can significantly improve the robustness of our model. After training, the CNN model was able to capture unique features from DCE images and segment breast tumors, after which the predictive model extracted and selected radiomic features to diagnose tumor subtypes. In addition, the automatically segmented tumor masks had even better diagnostic results in the radiomics model compared with the manually segmented ROIs, which also confirms the feasibility of the proposed model in practical applications.CONCLUSION
This study adopted nnUnet framework, TSHN and radiomics to diagnose breast tumors in DCE images. The superior performance in the external test cohort indicated the proposed model may be feasible for automatic diagnosis of TNBC with high accuracy in multicenter data.Acknowledgements
No acknowledgement found.References
1. Fan M, Zhang P, Wang Y, et al. Radiomic analysis of imaging heterogeneity in tumours and the surrounding parenchyma based on unsupervised decomposition of DCE-MRI for predicting molecular subtypes of breast cancer. European Radiology. 2019;29(8):4456-4467.
2. Park HJ, Lee SS, Park B, et al. Radiomics Analysis of Gadoxetic Acid-enhanced MRI for Staging Liver Fibrosis. Radiology. 2019;290(2):380-387.
3. Chen C, Ou X, Wang J, et al. Radiomics-Based Machine Learning in Differentiation Between Glioblastoma and Metastatic Brain Tumors. Frontiers in Oncology. 2019;278(1):82-94.
4. Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S. Liver Fibrosis: Deep Convolutional Neural Network for Staging by Using Gadoxetic Acid-enhanced Hepatobiliary Phase MR Images. Radiology. 2018;287(1):146-155.
5. Isensee F, Jaeger P F, Kohl S, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods. 2021;18(2):203-211.
6. Shinohara RT, Sweeney EM, Goldsmith J, et al. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clinical. 2014;6(C):9-19.
7. Lei Z, Zhimeng L, Ruimei C, et al. Deep-learning method for tumor segmentation in breast DCE-MRI. Imaging Informatics for Healthcare, Research, and Applications. San Diego: Proceedings of SPIE. 2019;109540F.1-109540F.6.
8. Mahmoudi SA, Larhmam MA, et al. MRI Breast Tumor Segmentation Using Different Encoder and Decoder CNN Architectures. Journal of Computers. 2019;8(3):52-63.
Figures
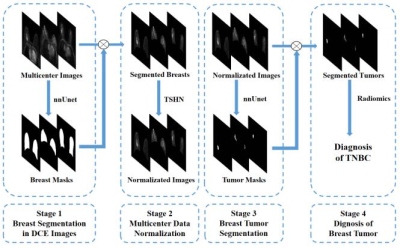
Figure 1. Overall framework of the proposed model, including a) breast segmentation in DCE images with nnUnet, and the segmented breast mask was used as the initial ROI for TSHN and tumor segmentation, b) multicenter data normalization using TSHN, c) breast tumor segmentation with nnUnet, and d) diagnosis of breast tumors based on radiomics analysis. (TSHN: Tissue-specific Histogram Normalization, TNBC: triple-negative breast cancer).
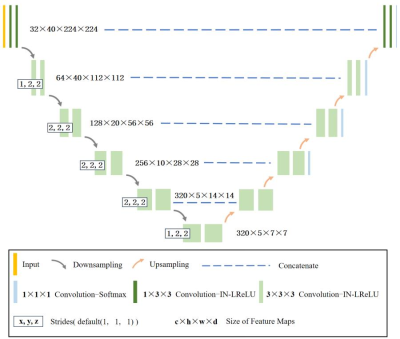
Figure 2. 3D full resolution U-Net architecture for tumor segmentation used in nnUnet framework.
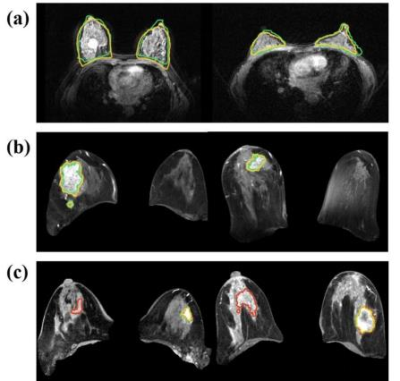
Figure 3. Examples of adopting nnUnet network to segment breast and tumors: a) breast segmentation, b) breast tumor segmentation, and c) comparison of segmentation results for models with and without TSHN normalization, the model with TSBH accurately predicted the tumor contours, while the model without TSBN showed large prediction errors. (Yellow contour: proposed automatic segmentation with TSHN, green contour: manually labeled mask, red contour: automatic segmentation without TSHN).
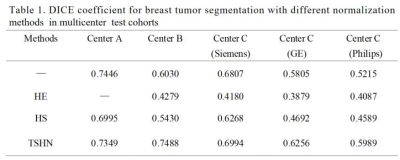
HE: histogram equalization, HS: histogram specification, TSHN: Tissue-specific Histogram Normalization
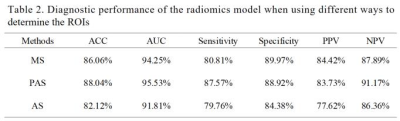
MS: manual segmentation, PAS: proposed automatic segmentation with TSHN, AS: automatic segmentation without TSHN, ACC: accuracy, AUC: areas under the curve, PPV: positive predictive value, NPV: negative predictive value.