0816
Sparse Annotation Deep Learning for Prostate Segmentation of Volumetric Magnetic Resonance Images1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2University of Chinese Academy of Sciences, Beijing, China, 3Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application, Guangzhou, China, 4Peng Cheng Laboratory, Shenzhen, China, 5Faculty of Mathematical and Computer Sciences, University of Gezira, Wad Madani, Sudan
Synopsis
Keywords: Segmentation, Prostate
Deep neural networks (DNNs) have achieved unprecedented performances in various medical image segmentation tasks. Nevertheless, DNN training requires a large amount of densely labeled data, which are labor-intensive and time-consuming to obtain. Here, we address the task of segmenting volumetric MR images using extremely sparse annotations, for which only the central slices are labeled manually. In our framework, two independent sets of pseudo labels are generated for unlabeled slices using self-supervised and semi-supervised learning methods. Boolean operation is adopted to achieve robust pseudo labels. Our approach can be very important in clinical applications to reduce manual effort on dataset construction.Introduction
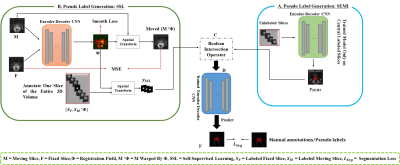
Prostate cancer is one of the most widespread categories of cancers in men, which makes accurate segmentation of prostate MR images extremely important for diagnosis and treatment [1], [2]. Manually segmenting 3D MR images by radiologists is a common but time-consuming and expensive practice. Consequently, automated algorithms with high performance are needed in real-life application scenarios. Deep neural networks have shown inspiring results in volumetric MR images segmentation. While DNNs perform well under the supervised learning paradigm [3], training the DNNs in this manner remains challenging since labeling 3D MR images is a tedious and costly process [4], [5]. It is difficult to obtain enough labeled data for DNN training. One key to decreasing the workload of manual labeling is making use of both labeled and unlabeled samples. Generating pseudo labels for the unlabeled data is one straightforward and effective strategy [6], [7]. When fully labeled data is not obtainable, there has generated a surge in interest in learning with unlabeled data. Unlike the current prevalent methods, which address the task of learning with limited densely labeled and large quantities of unlabeled data, we aim to tackle the challenge of sparse annotation learning when only the central slice of each 3D MR image is labeled manually. We propose a semi-supervised and self-supervised collaborative learning framework to generate pseudo labels for the unlabeled slices. Specifically, to achieve robust pseudo label generation, we use the semi-supervised learning method to progressively incorporate the unlabeled slices during the model training and pseudo label generation process, and a self-supervised learning technique to learn image deformations so as to transfer labels from the annotated central slice to the unlabeled slices. Then, we fuse the two pseudo label generation strategies by intersection operation to get superior pseudo labels for final segmentation network learning.Methodology
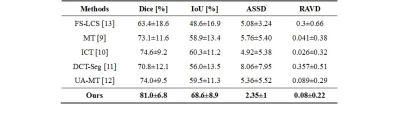
Figure 1 depicts our proposed semi-supervised and self-supervised collaborative learning framework. Inspired by [6], we propose a semi-supervised learning approach to integrate the unlabeled slices. A simple and straightforward method is designed, which makes use of the unlabeled slices by assigning unlabeled slices with network generated pseudo labels and then uses the pseudo labeled slices along with labeled slices to update the network. The model can iteratively enhance the quality of the pseudo labels since pseudo labeling is performed in an iterative fashion. The loss function adopted is the combination of dice loss and cross-entropy loss. To enhance the quality of self-generated pseudo labels, a self-supervised learning method is also proposed for pseudo label generation. It is based on image registration, which learns an image-level transformation field and transfer labels from the labeled central slice to the unlabeled slices utilizing the field. As a follow-up to our previous work [8], slice-based image registration is optimized using a deformable transformation framework. Finally, the semi-supervised learning and self-supervised learning methods are combined to obtain more accurate pseudo labels for the final target segmentation network optimization. In this case, an intersection is a good solution ($$$y_{final} = y_{SEMI} \cap y_{SSL}$$$). Eventually, the data with manual labels or self-generated pseudo labels are used to train a simple and general encoder-decoder segmentation network to segment the prostate regions in 3D MR images. This method is validated using the MICCAI Prostate MR Image Segmentation (PROMISE12) challenge dataset [1]. It consists of 50 3D MR images. 5-fold cross-validation experiments were conducted. Intersection-over-union (IoU), Dice score (Dice similarity coefficient, DSC), average symmetric surface distance (ASSD), and relative area/volume difference (RAVD) are calculated for quantitative evaluation. Better segmentation results are indicated by higher DSC and IoU values as well as lower RAVD and ASSD values.Results and Discussion
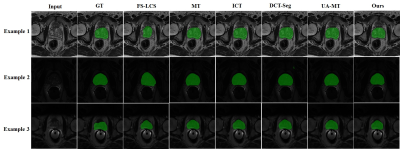
We compare our approach to four semi-supervised learning approaches (mean teacher (MT) [9], interpolation consistency training (ICT) [10], deep co-training (DCT-Seg) [11], and uncertainty-aware mean teacher (UA-MT) [12]) to assess the effectiveness of our proposed strategy. Table 1 reports the evaluation metrics of different approaches. It is noteworthy that our framework performs better on all evaluation metrics than the existing semi-supervised methods without implementing a complicated network architecture. It suggests that we can obtain rich information from the unlabeled slices using our proposed semi-supervised and self-supervised collaborative learning framework. Example prostate segmentation results are plotted in Figure 2. The segmentation maps of our proposed method have higher overlap ratios with the reference than those of other methods and yield fewer false positives, indicating better segmentation performance.Conclusion
In this paper, we introduced a semi-supervised and self-supervised collaborative learning framework for 3D prostate MR image segmentation with sparse annotation, for which the ground truth labels were provided for only one central slice of each 3D training data. Self-supervised learning and semi-supervised learning methods are proposed to generate two independent sets of pseudo labels. To obtain better pseudo labels for final segmentation network optimization, we combined the two sets of pseudo labels together using an intersection technique. Encouraging results are obtained by our framework on a public dataset for prostate segmentation. Our proposed framework can be very helpful in clinical applications when it is difficult to gather enough human resources for data collection and annotation.Acknowledgements
This research was partly supported by Scientific and Technical Innovation 2030-"New Generation Artificial Intelligence" Project (2020AAA0104100, 2020AAA0104105), the National Natural Science Foundation of China (61871371,62222118,U22A2040), Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application(No. 2022B1212010011), the Basic Research Program of Shenzhen (JCYJ20180507182400762), Shenzhen Science and Technology Program (Grant No. RCYX20210706092104034), AND Youth Innovation Promotion Association Program of Chinese Academy of Sciences (2019351).References
[1] G. Litjens, et al., “Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge,” Medical Image Analysis, vol. 18, no. 2, pp. 359-373, 2014.
[2] Osman, Y. B. M., Li, C., Xue, Z., Zheng, H., & Wang, S. “Accurate Prostate Segmentation in MR Images Guided by Semantic Flow”, in ISMRM, 2022.
[3] X. Chen, et al., “Recent advances and clinical applications of deep learning in medical image analysis,” Medical Image Analysis, vol. 79, pp. 102444, 2021.
[4] Umapathy, Lavanya, et al. "Learning to segment with limited annotations: Self-supervised pretraining with regression and contrastive loss in MRI", in ISMRM, 2022.
[5] A. Esteva, et al., “A guide to deep learning in healthcare,” Nature Medicine, vol. 25, pp. 24-29, 2019.
[6] D.-H. Lee, “Pseudo-label: the simple and efficient semi-supervised learning method for deep neural networks,” in ICML 2013 Workshop on Challenges in Representational Learning, vol. 3, 2013.
[7] S. Wang, et al., “Annotation-efficient deep learning for automatic medical image segmentation,” Nature Communications, vol. 12, pp. 5915, 2021.
[8] W. Huang, et al, “A coarse-to-fine deformable transformation framework for unsupervised multi-contrast MR image registration with dual consistency constraint,” IEEE Transactions on Medical Imaging, vol. 40, no. 10, pp. 2589-2599, 2021.
[9] A. Tarvainen, and H. Valpola, “Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results,” in NIPS, vol. 30, 2017.
[10] V. Verma, et al., “Interpolation consistency training for semi-supervised learning,” Neural Networks, vol. 145, pp. 90-106, 2022.
[11] J. Peng, et al., “Deep co-training for semi-supervised image segmentation,” Pattern Recognition, vol. 107, pp. 107269, 2020.
[12] L. Yu, et al., “Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation,” in MICCAI, pp. 605–613, 2019.
[13] T. Falk, et al., “U-Net: deep learning for cell counting, detection, and morphometry,” Nature Methods, vol. 16, no. 1, pp. 67-70, 2019.
Figures