0813
GIF_boost : A Generalisable Hybrid Brain Tissue Segmentation with DeepLearning1Centre for Medical Image Computing (CMIC), University College London, London, United Kingdom, 2Neuroradiological Academic Unit, UCL Queen Square Institute of Neurology, University College London, London, United Kingdom, 3Department of Radiology, Charité School of Medicine and University Hospital Berlin, Berlin, Germany, 4Radiology & Nuclear Medicine, VU University Medical Center, Amsterdam, Netherlands, 5e-Health Centre, Universitat Oberta de Catalunya, Barcelona, Spain
Synopsis
Keywords: Segmentation, Segmentation
Image segmentation and parcellation can provide quantitative assessment of the brain and can guide diagnosis and treatment decision-making. Geodesic Information Flow (GIF) is a freely available brain tissue segmentation and parcellation MRI-based tool using a classical label fusion approach. In this work, we introduce GIF_boost, a hybrid solution that takes advantages of deep learning to accelerate the bottleneck step of the template library registration. We compared GIF_boost with the original version of GIF and FreeSurfer (a state-of-the-art method). GIF_boost performed parcellation minimum 16 times faster. Parcellations had a similar Dice coefficient and Hausdorff distance and an improved volumetric quantification.
Introduction
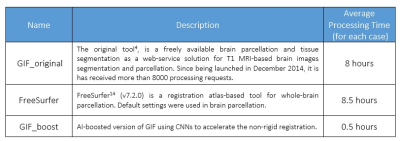
Longitudinal volumetric analysis of the brain can aid treatment planning and response monitoring in many neurological diseases1. Brain volume is obtained using segmentation and parcellation techniques1,6. Recently, deep learning networks such as ResNet and UNet1,11,13 have outperformed classical approaches in several segmentation challenges1,6,7. However, these algorithms are trained and largely assessed on challenge datasets of high-quality images, which may not be comparable to clinically acquired MRI data, so models may not be generalizable to other image acquisition protocols11,12.The GIF algorithm is a classical label fusion brain extraction and segmentation technique, which has been successfully tested on brain T1 scans from both hospital and open-source datasets. GIF is more generalizable because it is based on two steps, first a set of rigid and non-rigid registrations of the labelled template database to the input data using a modality-agnostic approach, and second, the label fusion of the results in the final segmentation and parcellation. The disadvantage of GIF is the long processing time of about 8 hours on a high-performance cluster4. This is due to the large number of rigid and non-rigid registrations for the whole template library. We aim to include deep learning to accelerate the registrations steps in the GIF pipeline.
Methods
Datasets and manual segmentationTo compare the performance of the chosen algorithms, we used 95 T1 healthy control manually delineated scans from the NeuroMorphometrics database including 30 OASIS3, 13 CANDI16, 30 ADNI17, 1 Colin2718, and 22 subjects who contributed their own scans. The NeuroMorphometrics brain parcellation delineates 208 brain regions and structures following the Desikan-Killiany-Tourville protocol.
Segmentation Algorithms
We compared our deep-learning-enhanced GIF_boost with the classic GIF implementation (GIF_original) and FreeSurfer14, a state-of-the-art parcellation method (Table 1).
Modifications
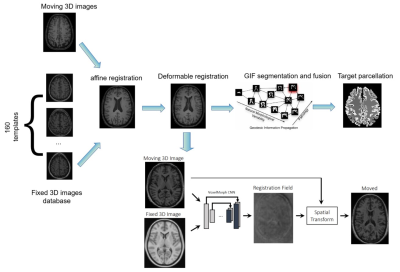
Fully AI-based brain segmentation methods are difficult to generalize to different input data, such as a lesioned, pathological brains or changes in image quality and modality11. Therefore, hybrid solutions can merge the advantages of each world (AI and non-AI), including specific steps for speeding up by AI solutions and keeping the non-generalizable steps performed by non-AI approaches.
Figure 1 provides an AI solution of GIF_boost, based on the VoxelMorph model2. VoxelMorph is based on a convolutional neural network. The VoxelMorph parameters were optimized on 3731 T1–weighted brain MRI scans from eight publicly available datasets (OASIS3, ABIDE5, ADHD2008, MCIC9, PPMI10, HABS15, Harvard GSP19, and FreeSurfer Buckner4014). For inference data, VoxelMorph rapidly computed a deformation field by directly evaluating the function parameterized by the convolutional network.
Evaluation
We quantitatively analysed the parcellation methods using the following metrics: “Dice” – Dice score coefficient, “HD” – Hausdorff distance and “Volume Diff” – volume difference in voxels between segmentation and ground truth. We evaluated 9 clinically relevant regions of interest: Brainstem, White Matter (WM), Cortical Grey Matter (CGM), and finally the right and left Hippocampus, Putamen and Thalamus. Bland-Altman plots have been used for the comparison between GIF versions.
Results
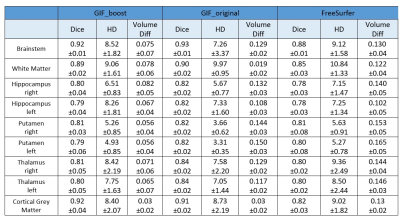
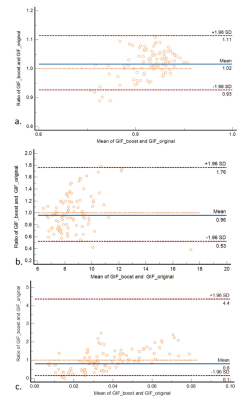
Results for the automated segmentation and parcellation of all testing data scans are presented in Table 2. In brain areas with relatively small volume, the Dice scores were lower using the GIF_boost model than GIF_original. However, there was no statistically significant difference between the Dice scores of both methods for brainstem, WM, and CGM tissue segmentations (p=0.053, 0.061, 0.083 respectively). The HD metric for brainstem, thalamus and CGM were not statistically significantly different between GIF models (p=0.145, 0.582, 0.176 respectively). However, the volume difference of all 9 regions computed with GIF_boost was smaller than using GIF_original, indicating that the GIF_boost segmentations were more accurate. On the other hand, FreeSurfer offer competitive results especially on volume difference, however it is slightly lower in Dice and HD compared to two GIF models. The computing time per case was reduced from 8 hours using GIF_original, and 8.5 hours with FreeSurfer to 0.5 hours using GIF_boost.CGM is hard to segment and is of clinical importance in parcellation1,6, Figure 2 presents a Bland-Altman CGM plot to compare the performance of the two GIF models. Figure 2a shows a higher Dice ratio (i.e. 1.02), meaning GIF_boost is outperforming GIF_original in CGM (p>0.05). Figure 2b shows the HD ratio of GIF_boost versus GIF_original is 0.96 (p>0.05). Finally, Figure 2c shows the Volume Diff with a mean ratio of 0.8, meaning GIF_boost is predicting more precise volumes in comparison to GIF_original (p<0.001).
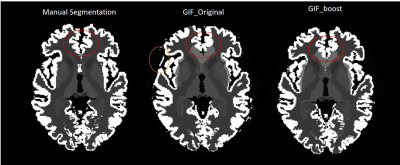
From a qualitative point-of-view, Figure 3, GIF_boost is providing more robust and less noisy parcellations on the brain borders (CGM), especially in the cingulate gyrus, and on the occipital part of the brain.
Discussion and Conclusion
In this study, we introduced an accelerated brain parcellation pipeline. The new method is a hybrid solution, where deep learning was used for accelerating the registration process of aligning the templates, while for the label fusion step, we used the classical approach. The new hybrid version obtained excellent performance in terms of the Dice and Hausdorff Distance. Moreover, it offered better results in terms of volume differences than the original model while it is minimum 16 times faster than before.Our findings suggest that the GIF tool, when enhanced with deep learning (GIF_boost), performs fast, accurate, and robust tissue segmentation and brain parcellation.
Acknowledgements
The GIF can be found at http://niftyweb.cs.ucl.ac.uk/program.php?p=GIF. FB, JW, GP, FP are supported by the NIHR Biomedical Research Centre at UCLH. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.References
1. Pereira, Sérgio, et al. Automatic brain tissue segmentation of multi-sequence MR images using random decision forests. Proceedings of the MICCAI grand challenge on MR brain image segmentation (MRBrainS’13) (2013).
2. Balakrishnan, Guha, et al. Voxelmorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging 38.8 (2019): 1788-1800.
3. D. S. Marcus, T. H. Wang, J. Parker, J. G. Csernansky, J. C. Morris, and R. L. Buckner, Open Access Series of Imaging Studies (OASIS): crosssectional MRI data in young, middle aged, nondemented, and demented older adults. Journal of cognitive neuroscience, (2007), vol. 19, no. 9, pp. 1498–1507.
4. Cardoso, M. Jorge, et al. Geodesic information flows: spatially-variant graphs and their application to segmentation and fusion. IEEE transactions on medical imaging 34.9 (2015): 1976-1988. 5. A. Di Martino, C.-G. Yan, Q. Li, E. Denio, F. X. Castellanos, K. Alaerts, J. S. Anderson, M. Assaf, S. Y. Bookheimer, M. Dapretto et al. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular psychiatry, vol. 19, no. 6, pp. 659–667, 2014 6. Ranta, Marin E., et al. Automated MRI parcellation of the frontal lobe. Human brain mapping 35.5 (2014): 2009-2026.
7. Akkus, Zeynettin, et al. Deep learning for brain MRI segmentation: state of the art and future directions. Journal of digital imaging 30.4 (2017): 449-459.
8. M. P. Milham, D. Fair, M. Mennes, S. H. Mostofsky et al. The ADHD200 consortium: a model to advance the translational potential of neuroimaging in clinical neuroscience. Frontiers in systems neuroscience, vol. 6, p. 62, 2012
9. R. L. Gollub, J. M. Shoemaker, M. D. King, T. White, S. Ehrlich, S. R. Sponheim, V. P. Clark, J. A. Turner, B. A. Mueller, V. Magnotta et al. The mcic collection: a shared repository of multi-modal, multisite brain image data from a clinical investigation of schizophrenia. Neuroinformatics, vol. 11, no. 3, pp. 367–388, 2013
10. K. Marek, D. Jennings, S. Lasch, A. Siderowf, C. Tanner, T. Simuni, C. Coffey, K. Kieburtz, E. Flagg, S. Chowdhury et al. The parkinson progression marker initiative (ppmi). Progress in neurobiology, vol. 95, no. 4, pp. 629–635, 2011.
11. Chen, Hao, et al. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage 170 (2018): 446-455.
12. Zhang, Xukun, et al. Confidence-Aware Cascaded Network for Fetal Brain Segmentation on MR Images. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2021.
13. Li, Suiyi, et al. Cascade dense-unet for prostate segmentation in MR images. International Conference on Intelligent Computing. Springer, Cham, 2019.
14. Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole Brain Segmentation: Automated Labeling of Neuroanatomical Structures in the Human Brain. Neuron. 2002 Jan 31;33(3):341–55.
15. A. Dagley, M. LaPoint, W. Huijbers, T. Hedden, D. G. McLaren, J. P. Chatwal, K. V. Papp, R. E. Amariglio, D. Blacker, D. M. Rentz et al. Harvard aging brain study: dataset and accessibility. NeuroImage, 2015.
16. Kennedy DN, Haselgrove C, Hodge SM, Rane PS, Makris N, Frazier JA. CANDIShare: a resource for pediatric neuroimaging data. Neuroinformatics. 2012;10(3):319-322. doi:10.1007/s12021-011-9133-y
17. Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201-209. doi:10.1212/WNL.0b013e3181cb3e25
18. Holmes CJ, Hoge R, Collins L, Woods R, Toga AW, Evans AC. Enhancement of MR images using registration for signal averaging. J Comput Assist Tomogr. 1998;22(2):324-333. doi:10.1097/00004728-199803000-00032
19. A. J. Holmes, M. O. Hollinshead, T. M. OKeefe, V. I. Petrov, G. R. Fariello, L. L. Wald, B. Fischl, B. R. Rosen, R. W. Mair, J. L. Roffman et al. Brain genomics superstruct project initial data release with structural, functional, and behavioral measures. Scientific data, vol. 2, 2015
Figures