0808
InnerEye as a Tool for Accurate Hippocampal Segmentation1Centre for Medical Image Computing, Department of Computer Science, University College London, London, United Kingdom, 2Lysholm Department of Neuroradiology, National Hospital for Neurology and Neurosurgery, University College London Hospitals NHS Foundation Trust, London, United Kingdom, 3Department of Brain Repair and Rehabilitation, UCL Institute of Neurology, University College London, London, United Kingdom, 4Department of Medical Physics & Biomedical Engineering, University College London, London, United Kingdom, 5Centre for Microscopy, Characterisation & Analysis, University of Western Australia, Perth, Australia, 6Health Futures, Microsoft Research Cambridge, Cambridge, United Kingdom, 7Department of Radiology and Nuclear Medicine, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam UMC, Amsterdam, Netherlands
Synopsis
Keywords: Segmentation, Brain, Hippocampus
Accurate hippocampal segmentation tools are critical for monitoring neurodegenerative disease progression on MRI and assessing the impact of interventional treatment. Here we present the InnerEye hippocampal segmentation model and evaluate this new model against three standard segmentation tools in an Alzheimer’s disease dataset. We found InnerEye performed best for Dice score, precision and Hausdorff distance. InnerEye performs consistently well across the different cognitive diagnoses, while performance for other methods decreased with cognitive decline.
Introduction
The hippocampus plays an important role in neurodegenerative diseases such as Alzheimer’s disease, where hippocampal volume provides an early biomarker of disease1. It is therefore important that we can accurately segment the hippocampus throughout the disease time-course, to monitor progression and to assess the impact of interventions, e.g. in clinical trials2. However, the hippocampus is a key region where openly available tools lack robustness, particularly in patients with conditions that affect hippocampal size and shape. While manual segmentation provides the gold-standard, it is time-consuming and costly, so there remains a clinical need for fast, accurate and robust automated segmentation tools.In this work, we present the InnerEye hippocampal segmentation model and evaluate it against various commonly used segmentation tools: FreeSurfer3, FastSurfer4 and HIPPOSEG5. In doing so, we compare the model to an atlas-based model (FreeSurfer), a deep learning model (FastSurfer), and a hippocampal-specific atlas-based model currently used in clinical practice at our institution (HIPPOSEG).
Methods
Data1155 T1 MP-RAGE MRI scans with corresponding ground truth hippocampal segmentations were obtained from the Alzheimer’s disease neuroimaging initiative (ADNI) database (adni.loni.usc.edu). These were collected from 484 subjects over separate visits. Scans had been pre-processed by ADNI using a combination of Gradwarp, B1-field correction and N3-bias correction. Ground truth segmentations6, provided by ADNI, were created semi-automatically using the Medtronic surgical navigation technology7.
Table 1 provides the data splits for training and validation of the InnerEye hippocampal model, and the test set used to evaluate all models. For each model, left and right hippocampi were analysed separately.
InnerEye Hippocampal Segmentation model
InnerEye is a toolbox for training and evaluating deep learning models on 3D medical images. It has shown success in, for example, segmentation models of structures in the head and pelvis for radiotherapy planning for head, neck and prostate cancers in CT scans8. We used InnerEye to generate a model for hippocampal segmentation of T1-weighted MRI (https://github.com/microsoft/InnerEye-DeepLearning/blob/main/docs/source/md/hippocampus_model.md), which we refer to simply as InnerEye in this abstract. The model consists of a 5-fold ensemble model of 3D U-nets.
Comparison Segmentation Tools
FreeSurfer3 (v7.2.0) is a registration, atlas-based model for whole-brain parcellation. FastSurfer4 (running on docker build gpu-v1.1.1) is a deep-learning model trained on the output of FreeSurfer (v6.0). Default settings were used in brain parcellation for both FastSurfer and FreeSurfer.
The HIPPOSEG5 analysis tool is a multi-atlas brain label fusion technique developed by the Centre for Medical Image Computing (CMIC) at UCL. We used the version of the tool developed to perform volumetry and relaxometry9 which has been used in clinical studies10,11 and is currently used clinically in the National Hospital for Neurology and Neurosurgery.
Models were evaluated using Dice score, Hausdorff distance, precision and recall.
Results
Figure 1 shows the performance of each model across different metrics. InnerEye outperforms all models in Dice, Hausdorff distance and precision. These results were significant (p<0.01) when assessing group differences using the standard t-test. The high precision of InnerEye segmentations across all subjects shows that InnerEye does not over-segment the hippocampus, in contrast to all other methods which show a low precision and an increase in Hausdorff distance. While InnerEye and FreeSurfer both perform well for recall, InnerEye shows a small number of outliers, which we do not observe in FreeSurfer.Figure 2 provides some qualitive assessment of each segmentation tool. Figure 2a shows the typical performance of each segmentation tool. InnerEye fits the ground truth segmentation well, while all other models over-segment the hippocampus. FreeSurfer shows the largest over-segmentation, while FastSurfer and HIPPOSEG over-segment the hippocampus at the head, and under-segment at the tail. Figure 2b shows a rare case where InnerEye has low recall, and FreeSurfer has high recall. Here InnerEye slightly under-segments the hippocampus. FreeSurfer captures the full segmentation, but also over-segments the model, resulting in a low Dice score.
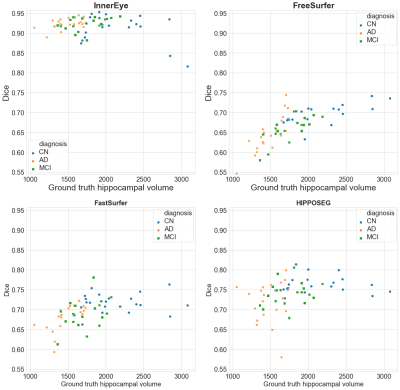
Figure 3 demonstrates how each model performs across cognitive diagnoses and different hippocampal volumes. FreeSurfer, FastSurfer and HIPPOSEG all perform worst for AD subjects, and best for CN. InnerEye performs consistently well across different hippocampal volumes and diagnoses, except for two CN outlier segmentations which show a lower Dice score (cf. Figure 2b). The poor performance of FreeSurfer, FastSurfer and HIPPOSEG at increased levels of atrophy strongly affects the accuracy of the hippocampal volume as a biomarker of AD.
Discussion and Conclusion
InnerEye significantly outperformed state-of-the art segmentation methods in the ADNI cohort and accurately segmented the hippocampus in a dataset of highly variable hippocampi, both in volume and topology.However, the InnerEye model was trained and tested on the same high-quality dataset from ADNI. While in our analysis we used a different subset of subjects then those used to train and validate the InnerEye model, the generalisability of InnerEye to other datasets needs to be assessed. In particular, the model will need to be tested on clinical datasets which may have lower quality images and/or different pre-processing steps. That said, InnerEye’s ability to segment the hippocampus accurately in cases and controls is a significant advance over what is currently freely available. Current work also focusses on estimating confidence of InnerEye output to detect failure cases automatically, which is important for embedding the tool in large-scale automated processing pipelines.
Acknowledgements
The InnerEye software is open source and can be found at https://github.com/microsoft/InnerEye-DeepLearning. FB JW, JST and TAY are supported by the NIHR Biomedical Research Centre at UCLH. Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.;Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.;Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
References
1. Jack CR, Petersen RC, Xu Y, O’Brien PC, Smith GE, Ivnik RJ, et al. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000 Aug 22;55(4):484–90.
2. Jack CR, Barkhof F, Bernstein MA, Cantillon M, Cole PE, DeCarli C, et al. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criteria for Alzheimer’s disease. Alzheimers Dement J Alzheimers Assoc. 2011 Jul;7(4):474-485.e4.
3. Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole Brain Segmentation: Automated Labeling of Neuroanatomical Structures in the Human Brain. Neuron. 2002 Jan 31;33(3):341–55.
4. Henschel L, Conjeti S, Estrada S, Diers K, Fischl B, Reuter M. FastSurfer - A fast and accurate deep learning based neuroimaging pipeline. NeuroImage. 2020 Oct 1;219:117012.
5. Winston GP, Cardoso MJ, Williams EJ, Burdett JL, Bartlett PA, Espak M, et al. Automated hippocampal segmentation in patients with epilepsy: Available free online. Epilepsia. 2013;54(12):2166–73.
6. Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, et al. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain. 2009 Apr;132(4):1067–77.
7. Hsu YY, Schuff N, Du AT, Mark K, Zhu X, Hardin D, et al. Comparison of automated and manual MRI volumetry of hippocampus in normal aging and dementia. J Magn Reson Imaging JMRI. 2002 Sep;16(3):305–10.
8. Oktay O, Nanavati J, Schwaighofer A, Carter D, Bristow M, Tanno R, et al. Evaluation of Deep Learning to Augment Image-Guided Radiotherapy for Head and Neck and Prostate Cancers. JAMA Netw Open. 2020 Nov 30;3(11):e2027426.
9. Vos SB, Winston GP, Goodkin O, Pemberton HG, Barkhof F, Prados F, et al. Hippocampal profiling: Localized magnetic resonance imaging volumetry and T2 relaxometry for hippocampal sclerosis. Epilepsia. 2020 Feb;61(2):297–309.
10. Caciagli L, Wandschneider B, Xiao F, Vollmar C, Centeno M, Vos SB, et al. Abnormal hippocampal structure and function in juvenile myoclonic epilepsy and unaffected siblings. Brain. 2019 Sep 1;142(9):2670–87.
11. Galovic M, Baudracco I, Wright-Goff E, Pillajo G, Nachev P, Wandschneider B, et al. Association of Piriform Cortex Resection With Surgical Outcomes in Patients With Temporal Lobe Epilepsy. JAMA Neurol. 2019 Jun 1;76(6):690–700.
Figures
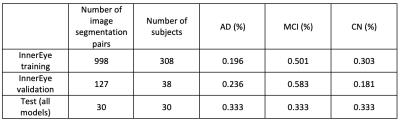
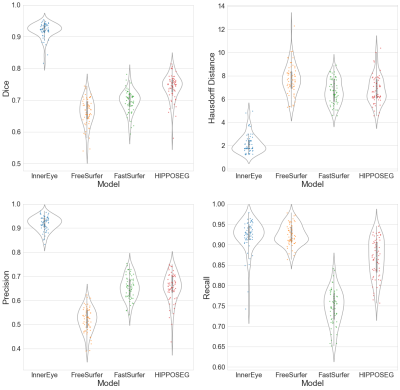
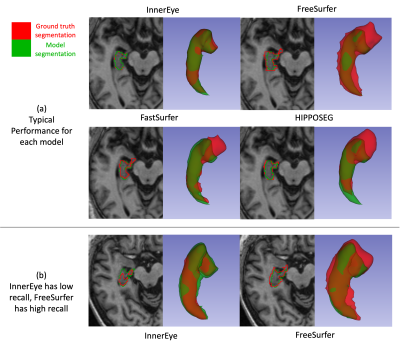
Figure 2: Here we show two cases with each model’s segmentation. (a) shows a case where each model has a typical performance (Dice scores are within the middle 50th percentile for each model’s Dice scores). Dice score for each segmentation: InnerEye=0.938, FreeSurfer=0.689, FastSurfer=0.694, HIPPOSEG=0.728. (b) shows a case where the InnerEye segmentation has a low recall, and FreeSurfer has a high recall. Dice and recall: InnerEye Dice=0.816, recall=0.742, FreeSurfer Dice=0.736, recall=0.921.