0806
Artifact-robust vascular segmentation for 3D phase-contrast MR angiography using a deep learning approach1Radiology, University of Wisconsin-Madison, Madison, WI, United States, 2Department of Radiology, Universität zu Lübeck, Lübeck, Germany, 3MIRAI Technology Institute, Shiseido Co., Ltd., Tokyo, Japan, 4Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 5Biomedical Engineering, University of Wisconsin-Madison, Madison, WI, United States, 6Medicine, University of Wisconsin-Madison, Madison, WI, United States, 7Emergency, University of Wisconsin-Madison, Madison, WI, United States
Synopsis
Keywords: Segmentation, Segmentation, phase contrast MRA
We developed a segmentation algorithm for PC-MRA using a deep-learning approach, with the goal of achieving artifact-robust segmentation for PC-MRA. To simulate flow-related artifacts of MRA, Gaussian noise and phase error were added to the k-space domain of the datasets. LadderNet consists of two consecutive U-nets with skip connections, and has been adopted as a training network for vessel segmentation. Retrospective studies demonstrated superior accuracy and precision of the proposed method over a conventional level set segmentation method.Introduction
Phase-contrast (PC) magnetic resonance angiography (MRA) can quantify and visualize blood flow in various body regions such as the brain1-3, heart4,5, and abdomen6,7. Furthermore, the derived complex difference (CD) images allow visualization of complex vessel morphology with excellent background signal suppression due to velocity encoding.Quantitative analysis of the acquired MR angiograms requires precise vessel segmentation. Several segmentation algorithms have been proposed for phase-contrast MRA8-12 as manual segmentation is cumbersome. Most conventional algorithms rely on the determination of an intensity threshold. However, intensity threshold-based methods are prone to error due to pulsation artifacts that are frequently observed in PC-MRA as ghosting of the vessel. Any high-signal artifacts included in the intensity-based segmentation will result in degraded segmentation quality.
Deep learning(DL)-based segmentation13 is emerging as a promising and more rapid segmentation strategy with improved performance and increased robustness to artifacts. In this study, we developed a segmentation algorithm for PC-MRA using a DL, with the goal to achieve artifact-robust segmentation for PC-MRA.
Methods
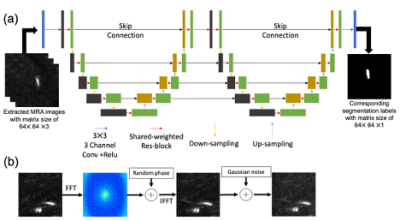
Ladder networkA LadderNet14 that consists of two consecutive U-net with skip connections was adopted as a training network for vessel segmentation of PC-MRA. Originally, the network was developed to demonstrate the segmentation of retina vessel images. In this study, we modified the network by using 3-channel 2D convolutions to utilize vessel structure information along the depth direction as shown in Figure 2(a).
Datasets
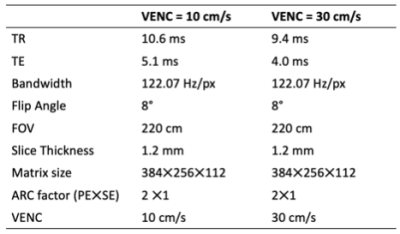
A set of 8 PC-MRA datasets were obtained from four volunteers recruited with IRB approval. Datasets from two subjects were used for training the network, and the rest was used for the test dataset. The PC-MRA images were acquired using a commercial 3D PC-MRA method (Inhance 3D Velocity, GE Healthcare, Waukesha, WI) on a 3.0T clinical MRI system (Signa Premier, GE Healthcare) and a 16-element head/neck coil. Acquisition parameters are summarized in Table 1.
Training for network
From the datasets, 47,742 patches were randomly extracted. The patches with background signal only were removed from the training datasets. The corresponding segmentation label images were used for the output of the network. As part of the training dataset, Gaussian noise and phase error15 were added in order to simulate noisy PC-MRA images with flow-related artifacts as shown in Figure 2(b).
The network was implemented in Python 3.8 using PyTorch. Adam optimizer with an initial learning rate of 0.001 was used for optimization of the network. A total of 100 epochs were used to train the network with a batch size of 16.
Manual Segmentation
Ground-truth datasets were generated with a semi-automatic segmentation pipeline using 3D Slicer, which is an open-source software platform16. Initial vessel segmentation results were extracted using a local threshold segmentation algorithm. After the segmentation, manual cleaning was performed to remove inappropriate regions.
Level-set segmentation
In comparison to the proposed method, we used an isosurface-initiated level-set segmentation as a benchmark. The segmentation was performed using the open-source vascular modeling tool kit software (VMTK)17. Parameters for iso-surface levels were determined to maximize the accuracy, as explained below, between the segmentation and ground-truth results.
Evaluation methods
Segmentation of the test datasets was implemented to compare segmentation performance of the proposed method. A slice-by-slice assessment of segmentation performance was performed using the accuracy and precision defined below,
$$Accuracy = \frac{TP+TN}{TP+FN+FP+TN},$$
$$Precision = \frac{TP}{TP+FP},$$
where TP, TN, FP, and FN denote true positive, true negative, false positive, and false negative, respectively.
Results
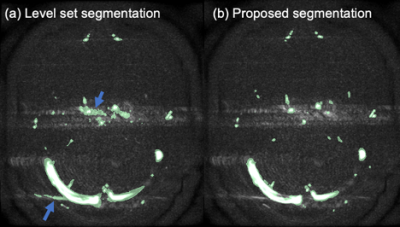
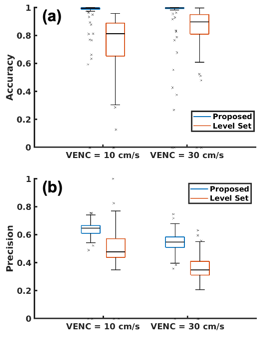
Figure 2 shows examples of extracted vessels using manual, level set, and proposed segmentation methods. Both Level set and the proposed method were capable of segmenting major blood vessels. Lebel set segmentation failed to label vessel voxels where the intensity was obscured or inhomogeneous due to pulsation artifacts as shown in Figure 3.Quantitative analysis also indicated that the proposed method (0.96±0.15 and 0.95±0.18 for VENC=10 and VEN=30cm/s) provides higher accuracy compared to level set segmentation (0.74±0.19 and 0.86±0.16 for VENC=10 and VENC=30cm/s) as shown in Figure 4(a). Also, the proposed methods achieved high precision (Figure 4(b)) segmentation, which means fewer false positive labeled voxels.
Discussion
In this study, we successfully developed a DL-based segmentation algorithm for PC-MRA that is insensitive to pulsation artifacts. To simulate noisy MRA images, Gaussian noise and phase error were added to the data. Retrospective studies demonstrated superior accuracy and precision of the proposed method over a conventional level set segmentation method.In MR angiograms with artifacts, conventional segmentation methods can be challenging since they rely heavily on intensities or seeds, which are susceptible to noise. In addition, it is often necessary to adjust parameters to obtain appropriate segmentation results. The proposed algorithm takes artifacts and noise into account and provides robust segmentation without parameter adjustments.
A major limitation of this study lies in the limited sample size. Although the training of the network was successful, a larger number of samples could be helpful to increase the robustness of the segmentation. Additionally, VENC may affect segmentation performance. To assess generalization performance, additional in vivo experiments should be conducted with different VENCs.
This study developed an artifact-robust segmentation algorithm using DL. In vivo and quantitative analyses indicated the robustness of the algorithm.
Data Availability
Source code of the network training and trained model are available at https://github.com/dtamadauw/PCMRA_Vessel_Segmentation_LadderNet.Acknowledgements
This work is supported by Shiseido Co., Ltd. We wish to acknowledge the support from GE Healthcare who provides research support to the University of Wisconsin. Dr. Reeder is the Fred Lee Sr. Endowed Chair of Radiology.References
1. Turski P, Scarano A, Hartman E, Clark Z, Schubert T, Rivera L, Wu Y, Wieben O, Johnson K. Neurovascular 4DFlow MRI (Phase Contrast MRA): emerging clinical applications. Neurovascular Imaging 2016;2(1):1-11.
2. Morgan AG, Thrippleton MJ, Wardlaw JM, Marshall I. 4D flow MRI for non-invasive measurement of blood flow in the brain: a systematic review. Journal of Cerebral Blood Flow & Metabolism 2021;41(2):206-218.
3. Youn SW, Lee J. From 2D to 4D Phase-Contrast MRI in the Neurovascular System: Will It Be a Quantum Jump or a Fancy Decoration? J Magn Reson Imaging 2022;55(2):347-372.
4. Lotz J, Meier C, Leppert A, Galanski M. Cardiovascular flow measurement with phase-contrast MR imaging: basic facts and implementation. Radiographics 2002;22(3):651-671.
5. Gorecka M, Bissell MM, Higgins DM, Garg P, Plein S, Greenwood JP. Rationale and clinical applications of 4D flow cardiovascular magnetic resonance in assessment of valvular heart disease: a comprehensive review. Journal of Cardiovascular Magnetic Resonance 2022;24(1):1-22.
6. Yzet T, Bouzerar R, Allart JD, Demuynck F, Legallais C, Robert B, Deramond H, Meyer ME, Baledent O. Hepatic vascular flow measurements by phase contrast MRI and doppler echography: a comparative and reproducibility study. J Magn Reson Imaging 2010;31(3):579-588.
7. Oechtering TH, Roberts GS, Panagiotopoulos N, Wieben O, Roldán-Alzate A, Reeder SB. Abdominal applications of quantitative 4D flow MRI. Abdominal Radiology 2021:1-22.
8. Chung AC, Noble JA, Summers P. Fusing speed and phase information for vascular segmentation of phase contrast MR angiograms. Med Image Anal 2002;6(2):109-128.
9. Chung AC, Noble JA, Summers P. Vascular segmentation of phase contrast magnetic resonance angiograms based on statistical mixture modeling and local phase coherence. IEEE Trans Med Imaging 2004;23(12):1490-1507.
10. Chen C, Zhou K, Guo X, Wang Z, Xiao R, Wang G. Cerebrovascular segmentation in phase-contrast magnetic resonance angiography by multi-feature fusion and vessel completion. Comput Med Imaging Graph 2022;98:102070.
11. Persson M, Solem JE, Markenroth K, Svensson J, Heyden A. Phase contrast MRI segmentation using velocity and intensity. 2005. Springer. p 119-130.
12. Hassan H, Farag AA. Cerebrovascular segmentation for MRA data using level sets. 2003. Elsevier. p 246-252.
13. Livne M, Rieger J, Aydin OU, Taha AA, Akay EM, Kossen T, Sobesky J, Kelleher JD, Hildebrand K, Frey D. A U-Net deep learning framework for high performance vessel segmentation in patients with cerebrovascular disease. Frontiers in neuroscience 2019;13:97.
14. Zhuang J, Multi-path networks based on U-Net for medical image segmentation. arXiv preprint arXiv: 181007810 2018.
15. Tamada D, Kromrey M-L, Ichikawa S, Onishi H, Motosugi U. Motion Artifact Reduction Using a Convolutional Neural Network for Dynamic Contrast Enhanced MR Imaging of the Liver. Magnetic Resonance in Medical Sciences 2020;19(1):64-76.
16. Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magnetic resonance imaging 2012;30(9):1323-1341.
17. Izzo R, Steinman D, Manini S, Antiga L. The vascular modeling toolkit: a Python library for the analysis of tubular structures in medical images. Journal of Open Source Software 2018;3(25):745.
Figures