0805
Thalamic segmentation methods in the characterization of Alzheimer's disease1University of Reading, Reading, United Kingdom, 2University of Massachusetts Chan Medical School, Worcester, MA, United States
Synopsis
Keywords: Segmentation, Segmentation
We present a systematic comparison of three state of the art thalamic segmentation methods for T1 MRI. Segmentation performance against Krauth-Morel atlas was quantified on 100 young healthy subjects and thalamic atrophy as a function of Alzheimer's disease status was characterized on 540 older subjects.Introduction
Thalamic nuclei segmentation from anatomical T1 and T2 MRI data has been hampered by lack of image contrast to delineate intrathalamic and whole thalamus boundaries. Most thalamic nuclei segmentation methods, to date, have been based on Diffusion Tensor Imaging (DTI), which is limited by the lack of anisotropy in the largely grey-matter dominant thalamus, and functional MRI, which is limited by poor spatial resolution and distortion of the underlying echoplanar acquisition. As a result, these methods do not resolve small structures such as lateral and medial geniculate nuclei (LGN/MGN), and the anteroventral (AV) nucleus, which are critical in sensory perception, cognition, and episodic memory.Recently, there has been a renewed interest in thalamic segmentation based on anatomical T1-weighted MRI, like the Freesurfer Bayesian inference [1], and the THOMAS multi-atlas [2] approaches. These methods use different thalamic nomenclatures and produce parcellations which differ qualitatively from each other despite claims of closeness to the Morel atlas, which is based on histological staining of post-mortem brains of healthy older adults. Here, we systematically evaluate algorithms for thalamic segmentation using Freesurfer, THOMAS, and a deep-learning variant of THOMAS. We used data from healthy younger adults in the Human Connectome Project (HCP) to compare segmentations against the Krauth-Morel atlas [3]. We analyzed data from older adults from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database to characterize thalamic atrophy as a function of disease status. We also assessed the accuracy of each of the three methods to predict Alzheimer’s disease status.
Methods
100 HCP subjects chosen at random and 540 subjects from the ADNI dataset (119 healthy controls (HC), 208 early minor cognitive impairment (EMCI), 116 late minor cognitive impairment (LMCI), and 97 Alzheimer disease (AD) see [4,5] for selection criteria) were segmented using (a) Freesurfer [1] (b) THOMAS modified for use with T1-weighed images using mutual information metrics for nonlinear registration and majority voting [5] and (c) THOMAS-CNN [6] which uses T1-weighted images to first synthesize white-matter nulled (WMn) MPRAGE images and then segment the synthetic WMn images using 2.5-D U-nets (Figure 1). Non-linear registration of nuclei in the Krauth-Morel atlas to T1 space was performed using ANTs [7]. Freesurfer and Krauth-Morel nuclei were combined to match the Morel nomenclature used by THOMAS and THOMAS-CNN. We used Dice coefficients to compare 10 segmented thalamic nuclei per hemisphere from each approach with that of Krauth-Morel. We also used ANCOVA and ROC analyses to characterize atrophy in ADNI subjects.Results
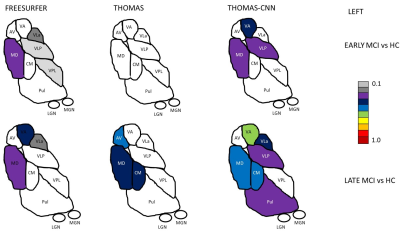
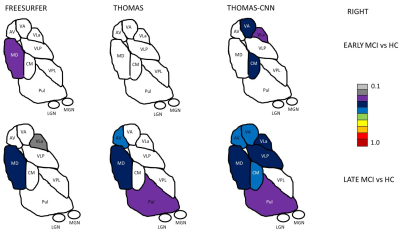
Two-way repeated measures ANOVAs found significant main effects of segmentation approach and hemisphere (left, right) and significant interactions of segmentation approach and hemisphere on Dice coefficients with Krauth-Morel for each nucleus (except for effects of hemisphere for VPL). Posthoc t-tests (Bonferroni corrected) are summarized in Figure 2 for left thalamic nuclei (similar results were found for the right hemisphere). THOMAS-CNN and Freesurfer approaches showed comparable performance (bilaterally, THOMAS-CNN had higher Dice for VLP, LGN, and Pul while Freesurfer had higher Dice for VA, CM and MDPf) and consistently outperformed THOMAS.Analysis of Covariance (ANCOVA) tests were used to assess if thalamic nuclei volumes for each segmentation approach differed between the 4 groups in the ADNI dataset after adjusting for age and intracranial volume; Dunnett’s test was used for pair-wise posthoc comparisons of significant ANCOVA models. ANCOVA results for each segmentation approach are summarized in Figure 3. Nuclei with significantly different pair-wise volumes for CN, EMCI, LMCI, and AD are presented separately for the left and right thalamus. Significant ANCOVAs with no pair-wise differences are denoted with an asterisk. Figure 4 shows thalamic nuclei atrophy colorized using Cohen’s d for the left side for HC-EMCI and HC-LMCI comparisons. The corresponding results for the right side are shown in Figure 5. The progression of atrophy from EMCI to LMCI is captured nicely by THOMAS-CNN with larger effect-sizes while Freesurfer does not exhibit a clear progression from EMCI to LMCI. THOMAS displays the progression but with reduced sensitivity and effect sizes.
An ROC analysis was performed using logistic regression to quantify the ability of each method to discriminate EMCI, LMCI, and AD from HC. AUC values for discrimination of AD and HC for Freesurfer, THOMAS, and THOMAS-CNN using all the individual thalamic nuclei volumes (adjusted for ICV/age) were 0.77, 0.81, and 0.85 respectively. AUC values for discrimination of LMCI and HC for Freesurfer, THOMAS, and THOMAS-CNN were 0.69, 0.73, and 0.76 respectively. Finally, AUC values for discrimination of EMCI and HC for Freesurfer, THOMAS, and THOMAS-CNN were 0.7, 0.69, and 0.73 respectively. Classification of HC-AD and HC-LMCI was more accurate using THOMAS-CNN than THOMAS and Freesurfer, while classification of HC-EMCI, was slightly better using THOMAS.
Conclusions
To our knowledge, this is the first work to systematically compare three recently published methods for thalamic nuclei segmentation using structural MRI in healthy younger and older adults. THOMAS-CNN showed the best accuracy in discriminating HC and AD and HC and LMCI as well as the largest effect sizes. The improved performance of THOMAS-CNN could be due to the fact it first synthesizes WMn, where intrathalamic contrast is heightened, prior to segmentation. Further work using manual segmentation is required to further compare these methods.Acknowledgements
No acknowledgement found.References
[1] Iglesias, J. E., Insausti, R., Lerma-Usabiaga, G., Bocchetta, M., Van Leemput, K., Greve, D. N., van der Kouwe, A., Alzheimer's Disease Neuroimaging Initiative, Fischl, B., Caballero-Gaudes, C., & Paz-Alonso, P. M. (2018). A probabilistic atlas of the human thalamic nuclei combining ex vivo MRI and histology. NeuroImage, 183, 314–326. https://doi.org/10.1016/j.neuroimage.2018.08.012
[2] Su, J. H., Thomas, F. T., Kasoff, W. S., Tourdias, T., Choi, E. Y., Rutt, B. K., & Saranathan, M. (2019). Thalamus Optimized Multi Atlas Segmentation (THOMAS): fast, fully automated segmentation of thalamic nuclei from structural MRI. NeuroImage, 194, 272–282. https://doi.org/10.1016/j.neuroimage.2019.03.021
[3] Krauth, A., Blanc, R., Poveda, A., Jeanmonod, D., Morel, A., & Székely, G. (2010). A mean three-dimensional atlas of the human thalamus: generation from multiple histological data. NeuroImage, 49(3), 2053–2062. https://doi.org/10.1016/j.neuroimage.2009.10.042
[4] Williams, B., Roesch, E., & Christakou, A. (2022). Systematic validation of an automated thalamic parcellation technique using anatomical data at 3T. NeuroImage, 258, 119340. https://doi.org/10.1016/j.neuroimage.2022.119340
[5] Bernstein, A. S., Rapcsak, S. Z., Hornberger, M., Saranathan, M., & Alzheimer’s Disease Neuroimaging Initiative (2021). Structural Changes in Thalamic Nuclei Across Prodromal and Clinical Alzheimer's Disease. Journal of Alzheimer's disease : JAD, 82(1), 361–371. https://doi.org/10.3233/JAD-201583
[6] Umapathy, L., Keerthivasan, M. B., Zahr, N. M., Bilgin, A., & Saranathan, M. (2022). Convolutional Neural Network Based Frameworks for Fast Automatic Segmentation of Thalamic Nuclei from Native and Synthesized Contrast Structural MRI. Neuroinformatics, 20(3), 651–664. https://doi.org/10.1007/s12021-021-09544-5
[7] Avants, B. B., Epstein, C. L., Grossman, M., & Gee, J. C. (2008). Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis, 12(1), 26–41. https://doi.org/10.1016/j.media.2007.06.004
Figures
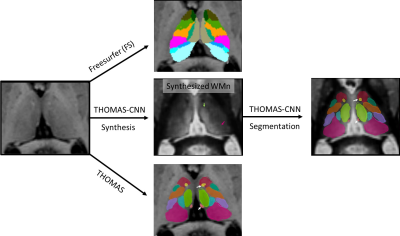
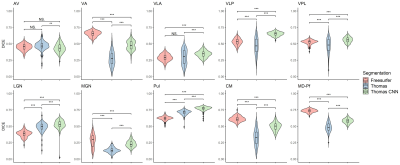

Figure 3: Pairwise analysis of Covariance (ANCOVA) results showing thalamic nuclei with significantly different volumes between healthy control (HC), early minor cognitive impairment (EMCI), late minor cognitive impairment (LMCI), and Alzheimer's disease (AD) individuals (after adjusting for age and intracranial volume) segmented using the Freesurfer [1], THOMAS [5] and THOMAS-CNN [6] approaches (Dunnett’s test). Significant ANCOVAs with no pair-wise differences are denoted with an asterisk.