0794
Motion Corrected DW-MRI With 3D Slice Level Motion Estimation In Pediatric Liver Tumors1RADIOLOGY, BOSTON CHILDREN'S HOSPITAL, HARVARD MEDICAL SCHOOL, BOSTON, MA, United States
Synopsis
Keywords: Liver, Motion Correction
Voxel misalignment due to unavoidable respiratory motion and bulk motion introduce large errors in DW-MRI quantitative parameter fitting. Apparent diffusion coefficient (ADC) is an effective tool for characterization of malignant tumors. In this study we evaluate the use of a motion correction method for DW-MRI imaging based on 3D slice level motion tracking using a rigid slice to volume registration and Kalman filtering. We show improvement in robustness of parameter estimation and reduction of blurring in b-value images for assessment of pediatric hepatoblastoma lesions.Introduction
Apparent diffusion coefficient (ADC) is an effective tool for detection and characterization of tumors due to sensitivity of DW-MRI parameters to tissue microstructure1. DW-MRI ADC is used in conjunction with contrast enhanced MRI to evaluate difficult diagnostic cases of malignant pediatric masses, such as hepatoblastoma2.Unavoidable respiratory motion and bulk motion introduces large errors in quantitative parameter fitting in DW-MRI. Averaging misaligned slices for improved SNR leads to blurring, and reduces lesion conspicuity. Fitted ADC has lower accuracy and robustness.
We use a retrospective motion correction method based on 3D slice level motion tracking with a 3D slice to volume registration for motion parameter estimation and Kalman filtering for motion tracking7-8. Our method relies on effective freezing of intra-slice motion in fast EPI acquisition. Thus, motion occurs only between acquired slices.
Methods
DW-MR images from 10 pediatric hepatoblasoma patients were acquired clinically at 3T (MAGNETOM Prisma, Siemens) using a free-breathing single-shot EPI:TR/TE=5500/59ms;matrix=156x120;FOV=312x240mm;slice=4mm;b-values=50,400,800 s/mm^2;6 diffusion gradient directions;1 repetition;acquisition-time= 4mins. Subsets of subjects were treated with chemotherapy, or ablation therapy. Images were retrospectively reviewed according to IRB protocol. In all cases a radiological report indicated large remaining lesions. All lesions were marked by a pediatric radiologist on the DW-MRI.Slice to volume alignment is defined as a set of contiguously acquired $$$N$$$ slices $$$s_{1}...s_{N}$$$ for which the coordinates are known only w.r.t. scanner orientation $$$\Omega_s$$$, and not to the reference anatomy $$$\Omega_a$$$. The goal is to find a rigid transformation matrix for each $$$s_{n}$$$, $$$T_{n}$$$, while constraining to some smoothness estimate w.r.t. (mostly) respiratory nature of abdominal motion. We pose this problem in reverse, assuming that a motion state $$$T_{n}$$$, that belongs to some trajectory $$$f\tau=f(T_{1..N})$$$, consists of $$$T_{n-1}+W{n-1}$$$, where $$$W_{n-1}$$$ is random patient movement. Such model can be represented with a Kalman filter, where the next update state is devised based on previous slice(s). We estimate $$$W_{n-1}$$$ by registering $$$s_{n}$$$ to a canonical volume $$$V_{0}$$$, which represents some reference space $$$\Omega_{c}$$$. $$$\Omega_{c}$$$ is updated every $$$k$$$ steps, as the b-value of the acquisition is changed.
Once all $$$T_{1..N}$$$ are estimated, scattered point cloud interpolation scheme is used to register all known data points to a regular meshgrid by weighting each data point’s distance to this grid via a Gaussian kernel. This motion estimation and reconstruction algorithm was written in C++ and is available on github.com/quin-med-harvard-edu/dSVRK, and as a docker image hub.docker.com/r/quinlab/dSVRK. Next, ADC was estimated on motion corrected and free-breathing (uncorrected) data using a linear least squares SVD approach in python.
To evaluate improvement in precision of estimated parameters after motion correction we used bootstrap analysis. Random removal of gradient directions in the original data was performed to generate subsampled data points at each bootstrap iteration. In total 61 iterations were performed. ADC model was repeatedly fit to each bootstrap-subsampled signal. Evaluation consisted of coefficient of variation (CoV) estimation of the ADC parameter over multiple bootstrap iterations.
Results
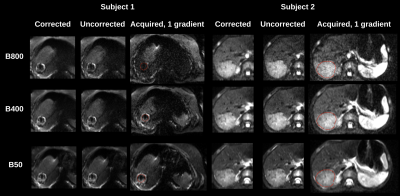
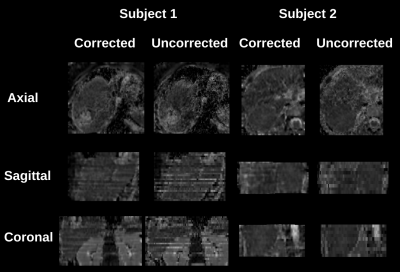
Figure 1 compares image quality of motion-corrected and free-breathing (uncorrected) b=50,400,800s/mm2 images of two hepatoblastoma subjects. Data is shown after geometrically averaging multiple gradient images at each b-value. Motion corrected data has improved image quality with sharper contour around the ablated tissue in Subject 1, and more details inside the lesion in Subject 2.Figure 2 shows severe motion effect on the ADC parameter maps in three orthogonal planes. While motion may appear less disturbing on the geometrically averaged b-value images in Figure 1, it is more pronounced on ADC maps. Uncorrected data yields strong stripy patterns that would bias any aggregate ADC measurements from a lesion or normal tissue.
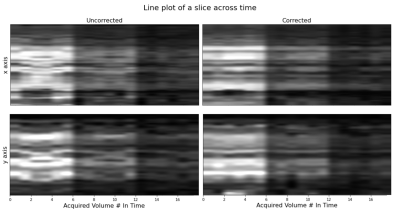
Figure 3 shows a line of voxels taken from coronal and axial planes plotted over time in the order of slice acquisition for all volumes and 3 b-values. The three distinct vertical stripes in the plot represent the changes in b-value over time (from b=50 to b=400, to b=800). In case of no motion, straight lines would appear across time. Motion corrected data exhibits improved alignment over time with more straight lines.
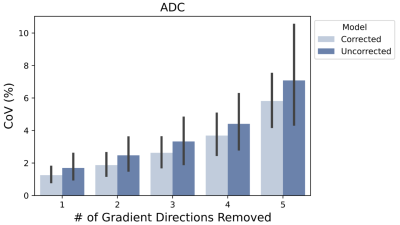
Figure 4 compares precision of ADC parameters from uncorrected and motion-corrected data over all subjects via bootstrap analysis. Motion corrected data exhibits consistently lower CoV (both mean and standard deviation over all subjects) indicating improved precision of ADC parameters . As expected, the CoV increases as more directions are removed from data, due to reducing SNR levels.
Conclusions and Discussion
Respiratory motion is an important contributor for poor accuracy and reproducibility in quantitative liver DW-MRI. The proposed motion correction method used 3D slice level parameter estimation, and regularisation of parameters over consecutively acquired slices with Kalman filtering. Results showed the effectiveness of this method in correcting the respiratory motion of the liver in DW-MR images. The motion corrected liver tumor regions had sharper contours with improved image quality and their ADC maps showed better spatial alignment and improved quality. Motion correction improved precision of ADC parameters by reducing their CoV% when removing one or multiple direction images from each b-value for bootstrap analysis. Despite the nonrigid nature of motion in the liver, a local 3D rigid motion correction method applied at the slice level was effective in correcting motion in DW-MRI of liver tumors.Acknowledgements
This work was supported in part by NIH grants R01 EB019483, R01 NS121657, R01 DK125561, R21 DK123569, R21 EB02962, and a pilot grant (PP-1905-34002) from the National Multiple Sclerosis Society.References
1. P. Caro-Domínguez et al. Can diffusion-weighted imaging distinguish between benign and malignant pediatric liver tumors? Pediatr Radiol. 2018;48:85–93.
2. Sodhi, Kushaljit Singh. "Diffusion-Weighted MRI in Children with Hepatoblastoma." Indian Journal of Pediatrics 89.10 (2022): 961-961.
3. G.R. Schooler, et al. Pediatric hepatoblastoma, hepatocellular carcinoma, and other hepatic neoplasms: consensus imaging recommendations from American College of Radiology Pediatric Liver Reporting and 4. Data System (LI-RADS) Working Group. Radiology 296.3 (2020): 493-497.
5. J.M. Guyader, et al. "Influence of image registration on apparent diffusion coefficient images computed from free‐breathing diffusion MR images of the abdomen." Journal of Magnetic Resonance Imaging 42.2 (2015): 315-330.
6. S. Kurugol, et. al. Motion-robust parameter estimation in abdominal diffusion-weighted MRI by simultaneous image registration and model estimation, Medical Image Analysis, 39, 124-132, 2017.
7. J. Zaffrani-Reznikov, et al. qDWI-Morph: Motion-compensated quantitative Diffusion-Weighted MRI analysis for fetal lung maturity assessment. arXiv preprint arXiv:2208.09836, 2022.
8. J. Coll‐Font, et al. Retrospective Distortion and Motion Correction for Free‐Breathing DW‐MRI of the Kidneys Using Dual‐Echo EPI and Slice‐to‐Volume Registration. Journal of Magnetic Resonance Imaging, 2020
9. S. Kurugol, et al. Motion-Robust Spatially Constrained Parameter Estimation in Renal Diffusion-Weighted MRI by 3D Motion Tracking and Correction of Sequential Slices, MICCAI RAMBO workshop, Lecture Notes in Computer Science, vol 10555. Springer, 2017.
Figures