0787
Multimodal neuroimaging of Col4A1-mutant mouse models of Gould Syndrome1UCSF, San Francisco, CA, United States
Synopsis
Keywords: Blood vessels, Rare disease
Gould syndrome is a multisystem disorder whose manifestations are highly variable. Animal studies suggest that allelic heterogeneity and genetic context contribute to the clinical variability. Clinical manifestations associated with Gould syndrome include blood–brain barrier (BBB) leakage, microbleeds, and white matter lesions. Here, we use multimodal MRI at 14.1Tesla to characterize radiological findings observed in mouse models of Gould syndrome caused by mutations in Collagen type IV alpha 1 (Col4a1). We show that multimodal MRI can successfully differentiate between disease subtypes based on anatomical changes as well as prevalence, number, volume and type of brain lesions.
Introduction
Collagen type IV alpha 1 and alpha 2 (COL4A1 and COL4A2) are major components of almost all basement membranes. COL4A1 and COL4A2 mutations cause a multisystem disorder that can affect any organ but typically involves the cerebral vasculature, eyes, kidneys and skeletal muscles. In recent years, patient advocacy and family support groups have united under the name of Gould syndrome1. The manifestations of Gould syndrome are highly variable, and animal studies suggest that allelic heterogeneity and genetic context contribute to the clinical variability. In this study, we used five different Col4a1 mutant mouse strains that recapitulate the clinical spectrum of Gould syndrome manifestations to investigate whether clinically relevant multimodal MR imaging could be used to differentiate between cerebrovascular disease subtypes that might benefit from distinct therapeutic interventions.Material & Methods
Animals: An allelic series of five Col4a1 mutations in mice (n=25 total, male only, age-matched, 51.2-53.7 weeks-old) (Fig.1A). These mutations consist in four distinct glycine missense mutations in the triple-helix-forming domain: G658D (n=5), G912V (n=5), G1038S (n=5), G1180D (n=5), and one missense mutation in the NC1 domain: S1582P (n=5) (Fig.1B). Five age-matched wild-type (WT) littermates mice were used as controls. A significant reduction in body weight was observed in mice with the G912V, G1038S and G1180D mutations (**p=0.002, *p=0.034 *p=0.0169, resp, Fig.1C).MR Acquisitions: Each imaging session was performed on a 14.1Tesla Agilent® system using a 1H Agilent® volume coil (inner diameter 40mm). All mice were imaged under 1.3-1.8% isoflurane using 4 optimized sequences of matching geometry (Field of view (FOV)=20x20mm2, matrix=256x256, 16 slices, 0.4mm slice thickness, 0.1mm interslice gap), for an overall acquisition time of ~40 min/animal. All sequence parameters are described in Fig.1.D.
Data Analysis: For brain tissue and lesion segmentation, a U-net Convolutional Neural Network2 algorithm was optimized in MATLAB® (Fig.2.A). One U-net was implemented for a skull-stripping operation that can be performed independent of MRI contrast (T2W, T1W, and SWI). Another U-net was trained for semantic segmentation of a specific brain tissue type (ventricle, etc.) or lesion type (hypointensity, etc). The training dataset came from published data from our group3. The ground truth for semantic segmentation was manually labelled in ImageJ. Both U-net architectures were trained with a 60/30/10 training/validation/testing split (skull-stripping: 192/96/32 T2w, T1w, and SWI slices; ventricle-segmenting: 48/24/8 T2w slices), encoder depth of 5, a Dice coefficient loss function and stochastic gradient descent with momentum (SGDM) optimization (Fig.2.B). Brain and ventricle volume were extracted from U-net and calculated for each animal. Ventricle-to-brain ratio was also evaluated. Lesions were extracted manually on SWI images (U-net will be used in the future), and the prevalence, number and volume of lesions were compiled. For all parameters, a 2-way ANOVA corrected for multiple comparisons was performed (*p<0.05, **p<0.01).
Results & Discussion
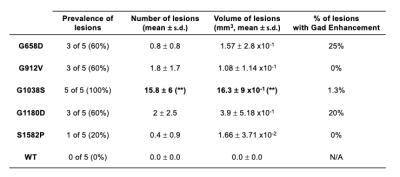
Representative brain images of all MRI modalities are shown for all five genotypes (Fig.3). and visually demonstrate how allelic heterogeneity leads to distinct radiological manifestations. As shown in Fig.4A, whole brain volume was significantly lower in G912V mice compared to WT controls (*p<0.05). In contrast, ventricle volume as well as ventricle-to-brain ratios were significantly higher in G658D and G1038S mice compared to WT controls (calculated by U-net, *p<0.05, Fig.4B&C). These results indicate that Col4a1 mutations in mice lead to large structural changes that are dependent upon the type of missense mutation.Looking at cerebral lesions, our results show that, as expected, SWI detected a higher number of hypointense lesions than T2W imaging (including smaller lesions) with sharper boundary (data not shown). Analysis of the prevalence, number, volume, and type of cerebral lesions was thus done on SWI data and is shown in Fig.5 for all 5 genotypes. The prevalence of the SWI-positive lesions dramatically varied from one genotype to another, from a 20% prevalence for S1582P mice to a 100% prevalence in G1038S mice. The number of lesions was significantly elevated compared to controls for G1038D (15.8±6 lesions per animal, **p<0.01). For the other 4 genotypes, lesions were detected, although statistical significance was not reached due to the lower prevalence. Similarly, the volume of lesions was significantly elevated in G1038S animals compared to controls (16.3±9 mm3, **p<0.01), while statistical significance was not reached for other genotypes due to lower prevalence. Finally, we also investigated the percentage of SWI-positive lesions presenting Gadolinium enhancement as detected by T1WI. Interestingly, these percentages were also very variable between genotypes, ranging from no enhancing lesions in S1582P and G912V mice, to 20% and 25% in G1180D and G658D mice, respectively. Notably, in G1038S mice, the genotype presenting the higher number of lesions, only 1.3% of them were enhancing, likely suggesting that this mutation has a more rapid time course than others, with active lesions present at earlier age.
Conclusions & Perspectives
Using a multimodal imaging approach, we showed that Col4a1 mutant mice recapitulate the radiological features commonly seen in individuals with Gould syndrome and that allelic heterogeneity influence the prevalence, number, volume and type of lesions. Immunohistochemistry studies are underway to determine the pathological signature of the lesions detected by multi-modality MRI. In the future, longitudinal imaging studies will be performed to characterize how allelic heterogeneity influences disease progression in these models.Acknowledgements
This work was supported by research grants NIH R61NS115132 and NIH RF1NS110044.References
1. Mao Mao, Tanav Popli, Marion Jeanne, Kendall Hoff, Saunak Sen, Douglas B. Gould; Identification of fibronectin 1 as a candidate genetic modifier in a Col4a1 mutant mouse model of Gould syndrome. Dis Model Mech 1 April 2021; 14 (4): dmm048231.
2. Ronneberger, O., Fischer, P., Brox, T. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab, N., Hornegger, J., Wells, W., Frangi, A. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. MICCAI 2015. Lecture Notes in Computer Science(), vol 9351. Springer, Cham.
3. Evan Yamasaki, Pratish Thakore, Sher Ali, Alfredo S. Solano, Xiaowei Wang, Xiao Gao, Cassandre Labelle-Dumais, Myriam M. Chaumeil, Douglas B. Gould, Scott Earley; Defective Ca2+-dependent activation of TRPM4 channels contribute to age-related cerebral small vessel disease in Col4a1 mutant mice. (Submitted to The Journal of Clinical Investigation)
Figures
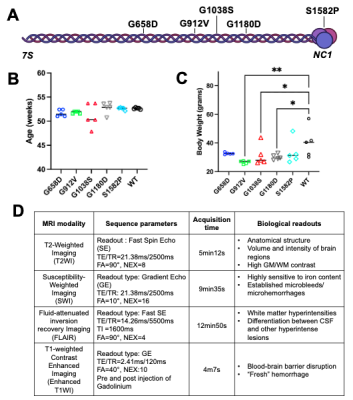
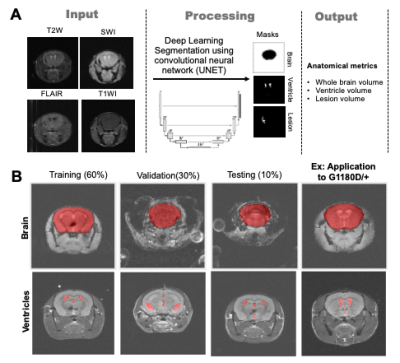
Figure 2. Volumetric analysis using U-net. (A) Two types of U-net are implemented: 1) skull-stripping U-net handling different MRI modality inputs; 2) tissue/lesion-specific U-net utilizing certain feature-weighted MRI contrast, such as T2W for ventricle masking. (B) The representative segmentation performance of skull-stripping and ventricle-segmenting U-net, where the training/validation/testing results come from a typical Gould syndrome genotype from elsewhere and the application testing is run on genotype G1180D/+.
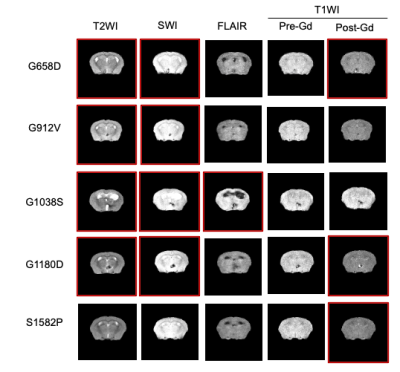
Figure 3. Multimodal MRI at 14.1Tesla. Representative images of all five different MRI modalities are shown for all five genotypes. Red boxes indicates positive findings.
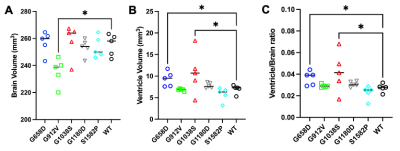
Figure 4. Brain and ventricle volumes, and ventricle-to-brain ratios in mouse models of Gould syndrome (A) Brain volume was significantly lower in G912V mice compared to WT controls (*p<0.05). (B) Ventricle volume was significantly higher in G658D and G1038S mice compared to WT controls (*p<0.05). (C) Ventricle-to-brain ratio was significantly higher in G658D and G1038S mice compared to WT controls (*p<0.05).