0711
A unified deep learning model for simultaneous cardiac cine MRI reconstruction, motion estimation and segmentation.1School of Biomedical Engineering, ShanghaiTech University, Shanghai, China, 2Shanghai Clinical Research and Trial Center, Shanghai, China, 3Department of Computer Science and Technology, University of Cambridge, Cambridge, United Kingdom
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence
Various deep learning methods have been proposed for cardiac cine MRI, including accelerated image reconstruction, cardiac motion estimation and segmentation, which are traditionally considered as separate tasks without exploiting the inter-task correlation. In this study, we propose a unified deep learning model to perform accelerated cine image reconstruction, motion estimation and segmentation simultaneously in an iterative framework, where correlations between tasks are exploited by compensating motion in reconstruction, semi-supervising segmentation using pseudo-labels generated by motion and improving motion estimation using intermediately reconstructed images. Experiment results show that the multi-task model outperformed single-task networks.
Introduction
Due to the wide application of cardiac cine MRI, numerous deep learning models have been proposed for cine image reconstruction, cardiac motion estimation and left ventricle (LV) and right ventricle (RV) segmentations. However, the reconstruction task and the post-processing tasks are usually considered separately. Several studies have shown that these tasks are closely related and benefit each other when considered in a multi-task model1–4. Previous studies have only tackled two tasks, such as joint motion estimation and segmentation1,4 and joint motion estimation and reconstruction2,3. In this study, we investigated the feasibility of a unified deep learning model to deal with the three tasks of accelerated cine reconstruction, motion estimation and segmentation simultaneously. A cascaded encoder-decoder network was adopted, where the performance of shared encoder and separated encoder for different tasks was investigated in designing the multi-task model. The multi-task model was also compared with the corresponding single-task networks.Methods
The proposed unified model is presented in Figure 1. Given $$$n_T$$$ undersampled cine MR image $$$X^{(0)}= \left\{x^{(0)}_1,\dots,x^{(0)}_{n_T}\right\}$$$ and initial zero motion as input, the network generates reconstructed cine images $$$X^{(i)}=\left\{ x^{(i)}_1,\dots,x^{(i)}_{n_T} \right\}$$$, predicted segmentation mask $$$\hat{S}^{(i)}$$$ and estimated velocity fields5 $$$\mathcal{V}^{(i)}= \left\{v^{(i)}_1,\dots,v^{(i)}_{n_T} \right\}$$$ for the $$$i$$$-th iteration, and $$$X^{(n)},\hat{S}^{(n)}, \mathcal{V}^{(n)}$$$ in the $$$n$$$-th iteration are the final outputs of the cascaded network with $$$n$$$ iterations.Diffeomorphic Groupwise Motion Estimation: We adopt a diffeomorphic group-wise motion estimation method to predict a set of velocity fields $$$\mathcal{V}^{(i)}=\left\{ v^{(i)}_1,\dots,v^{(i)}_{n_T}\right\}$$$ pointing to the geometry center of $$$X^{(i)}$$$, subsequently a pair of inversible transformations $$$\mathcal{T}^{(i)}=\left\{ T^{(i)}_1,\dots,T^{(i)}_{n_T}\right\}, {\mathcal{T}^{-1}}^{(i)}=\left\{ {T^{-1}}^{(i)}_1,\dots,{T^{-1}}^{(i)}_{n_T}\right\}$$$ are constructed by integrating from $$$\mathcal{V}^{(i)}$$$6. The template is the average of the warped dynamic images: $$${\bar{X}}^{\left(i\right)}=\frac{1}{n_T}\sum_{i=1}^{n_T}{T_i^{\left(i\right)}\circ X_i^{\left(i\right)}}$$$, where $$$\circ$$$ is the warping operator. The motion estimation task is trained in a self-supervised loss based on fully sampled images $$$X=\left\{ x_1,\dots,x_{n_T}\right\}$$$,
$$
\mathcal{L}_\mathrm{reg}^{(i)}=\frac{1}{n_T}\sum_{i=1}^{n_T}\left({\left\|\bar{X}-T_i^{(i)}\circ x_i\right\|}^2_2\right)+\frac{1}{n_T}\sum_{i=1}^{n_T}\left({\left\|x_i-{T^{-1}}_i^{(i)}\circ\bar{X}\right\|}^2_2\right)+\alpha\frac{1}{n_T}\sum_{i=1}^{n_T}\left({\left\|\nabla_\mathrm{xy}v_i^{(i)}\right\|}^2_2\right)+\beta\frac{1}{n_T}\sum_{i=1}^{n_T}\left({\left\|\nabla_\mathrm{t}v_i^{(i)}\right\|}^2_2\right)
$$
$$$\nabla_\mathrm{xy} v_i$$$ and $$$\nabla_\mathrm{t} v_i$$$ are gradients of the velocity fields $$$v_i$$$ in the spatial and temporal dimensions, $$$\alpha$$$ and $$$\beta$$$ are two hyper-parameters for constraining the spatial and temporal smoothness of the velocity fields.
Motion Augmented Reconstruction: Estimated motion can be used to aid in reconstruction task by fusing temporal information through motion2,7. Briefly, undersampled or intermediately reconstructed cine images are warped to the template with $$$\mathcal{T}^{(i)}$$$ and the template image is warped to the dynamic domain with $$${\mathcal{T}^{-1}}^{(i)}$$$, generating a set of motion-augmented dynamic images, where each frame contains information of all frames and is thus less undersampled. Data-consistency (DC) operation is then performed on the motion-augmented images: $$$X_\mathrm{aug}^{(i)}=\mathrm{DC} \left( {\mathcal{T}^{-1}}^{(i)} \circ \bar{X}^{(i)} \right)$$$. For $$$i$$$-th iteration, the network takes the concatenation of $$$X_\mathrm{aug}^{(i)}, X_\mathrm{aug}^{(0)}$$$ and $$$X^{(i-1)}$$$ as input and outputs $$$X^{(i)}$$$ as the reconstruction. The reconstruction loss function at the $$$i$$$-th iteration is:
$$
\mathcal{L}_\mathrm{rec}^{(i)}=\frac{1}{n_T}{\left \|X-X^{(i)}\right \|}^2_2
$$
Semi-supervised Segmentation: Cardiac cine MR images are commonly only annotated for the end-diastolic (ED) and end-systolic (ES) frames, resulting in manual labels $$$S=\left\{S_\mathrm{ES},S_\mathrm{ED}\right\}$$$. To exploit the dynamic information and segment all frames simultaneously, we generated pseudo labels with estimated motion: $$${\mathcal{T}^{-1}}^{\left(n\right)}\circ T_i^{\left(n\right)}\circ S_i,\;i\in\left\{i_\mathrm{ED},i_\mathrm{ES}\right\}$$$1,4. The segmentation loss is the weighted combination of the loss calculated with the manual labels and the generated labels:
$$
\mathcal{L}_\mathrm{edes}^{\left(i\right)}=-\sum_{i\in\left\{i_\mathrm{ED},i_\mathrm{ES}\right\}}{S_i\log{\left({\hat{S}}_i^{\left(i\right)}\right)}}
$$
$$
\mathcal{L}_\mathrm{warped}^{\left(i\right)}=-\sum_{i\in\left\{i_\mathrm{ED},i_\mathrm{ES}\right\}}\sum_{j\in\left\{1,\cdots,n_T\right\}}{\left({T_j^{-1}}^{\left(n\right)}\circ T_i^{\left(n\right)}\circ S_i\right)\log{\left({\hat{S}}_j^{\left(i\right)}\right)}}
$$
The network was trained in an end-to-end fashion with a composite loss function:
$$
\mathcal{L}=\sum_{i=1}^{n}2^{i-n}\left(\lambda_\mathrm{rec}\mathcal{L}_\mathrm{rec}^{\left(i\right)}+\lambda_\mathrm{reg}\mathcal{L}_\mathrm{reg}^{\left(i\right)}+\lambda_\mathrm{edes}\mathcal{L}_\mathrm{edes}^{\left(i\right)}+\lambda_\mathrm{warped}\mathcal{L}_\mathrm{warped}^{\left(i\right)}\right)
$$
where $$$\lambda_\mathrm{rec},\lambda_\mathrm{reg},\lambda_\mathrm{edes},\lambda_\mathrm{warped}$$$ are regularization parameters for each loss term.
Evaluation: The multi-task network was implemented using the 2D U-Net8 with recurrent unit to exploit the dynamic information, where two designs were considered: separate encoders for each task and a single encoder shared for all tasks. The considered baselines were the reconstruction and motion estimation networks trained separately with the same U-Net architecture to that in the multi-task model. The hyper-parameters in the multi-task model were manually tuned as $$$\alpha={10}^{-3},\beta={10}^{-4},\lambda_\mathrm{rec}={10}^2,\lambda_\mathrm{reg}={10}^3,\lambda_\mathrm{edes}=2,\lambda_\mathrm{warped}=1$$$. Experiments are performed in the ACDC9 dataset with simulated phase10 and 8-fold undersampling.
Results
The quantitative metrics are summarized in Table 1. The multi-task model with separate encoders had the best performance for almost all the tasks. Visual comparison of multi-task and single-task modes is illustrated in Figure 2,3,4 respectively for reconstruction, motion estimation and segmentation. The results indicate that our approach had accurate segmentation from highly undersampled images, and achieved higher PSNR and SSIM in both reconstruction and motion estimation than single-task methods, demonstrating the effectiveness of the multi-task model. The multi-task model achieved accurate segmentations over all cardiac phases, though only the labels at ED and ES frames were available for training, indicating the benefit of motion-warped labels. The multi-task model with shared encoder performed slightly worse than the counterpart with separate encoders.Discussion
We proposed a unified model to simultaneously perform accelerated reconstruction, segmentation and motion estimation in cardiac cine MR images by leveraging task correlations in a cascaded network. Our experiments show that the multi-task model significantly outperformed single-task reconstruction and motion estimation methods, which can be attributed to the motion augmented reconstruction and updating estimated motion with iteratively reconstructed images1,2. The shared encoder performed slightly worse than the separate encoders in the multi-task framework, indicating the simple way of enforcing shared features between tasks is not viable. In the future, more effective inter-task feature sharing method will be explored for a more efficient unified model.Acknowledgements
No acknowledgement found.References
1. Qin, C. et al. Joint Learning of Motion Estimation and Segmentation for Cardiac MR Image Sequences. in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018 (eds. Frangi, A. F., Schnabel, J. A., Davatzikos, C., Alberola-López, C. & Fichtinger, G.) 472–480 (Springer International Publishing, 2018).2. Yang, J., Küstner, T., Hu, P., Liò, P. & Qi, H. End-to-End Deep Learning of Non-rigid Groupwise Registration and Reconstruction of Dynamic MRI. Front. Cardiovasc. Med. 9, 880186 (2022).
3. Qi, H. et al. End-to-end deep learning nonrigid motion-corrected reconstruction for highly accelerated free-breathing coronary MRA. Magn. Reson. Med. 86, 1983–1996 (2021).
4. Qian, P., Yang, J., Lió, P., Hu, P. & Qi, H. Joint Group-Wise Motion Estimation and Segmentation of Cardiac Cine MR Images Using Recurrent U-Net. in Annual Conference on Medical Image Understanding and Analysis 65–74 (Springer, 2022).
5. Ashburner, J. A fast diffeomorphic image registration algorithm. NeuroImage 38, 95–113 (2007).
6. Dalca, A. V., Balakrishnan, G., Guttag, J. & Sabuncu, M. R. Unsupervised learning of probabilistic diffeomorphic registration for images and surfaces. Med. Image Anal. 57, 226–236 (2019).
7. Seegoolam, G. et al. Exploiting Motion for Deep Learning Reconstruction of Extremely-Undersampled Dynamic MRI. in Medical Image Computing and Computer Assisted Intervention – MICCAI 2019 (eds. Shen, D. et al.) 704–712 (Springer International Publishing, 2019)
8. Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. in International Conference on Medical image computing and computer-assisted intervention 234–241 (Springer, 2015).
9. Bernard, O. et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 37, 2514–2525 (2018).
10. Guerquin-Kern, M., Lejeune, L., Pruessmann, K. P. & Unser, M. Realistic Analytical Phantoms for Parallel Magnetic Resonance Imaging. IEEE Trans. Med. Imaging 31, 626–636 (2012).
Figures
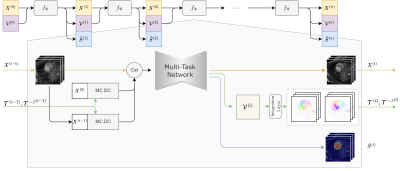
Figure 1. The proposed unified model iteratively optimizes reconstruction, motion estimation and segmentation in a cascaded network. In each iteration, network takes the concatenation of reconstructed images of the last iteration![]() and motion augmented images as input, generating updated reconstructions, segmentations and velocity fields.
and motion augmented images as input, generating updated reconstructions, segmentations and velocity fields.
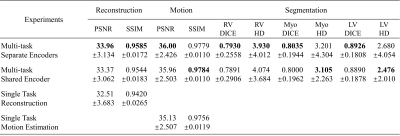
Table 1. Quantitative metrics for evaluating the reconstruction, motion estimation and segmentation performance. PSNR and SSIM were used for measuring the similarity between reconstructed images and fully sampled images. The similarity metrics were also evaluated between warped images and the template image to evaluate the motion estimation performance. Dice coefficient (DICE) and Hausdorff distance (HD) were evaluated using annotated ground truth on ED and ES frames.
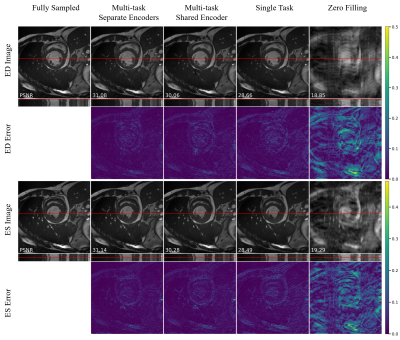
Figure 2. Visualization of reconstruction on an exemplary subject at ED and ES frames. Error map depicts the absolute error between fully sampled images and reconstructions.
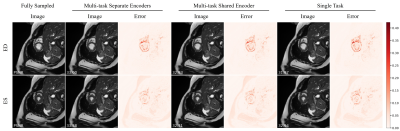
Figure 3. Visualization of generated cardiac cine images from estimated motions on an exemplary subject at ED and ES frames. Error maps depict the difference between the generated images and the original images, reflecting the accuracy of the estimated motion.
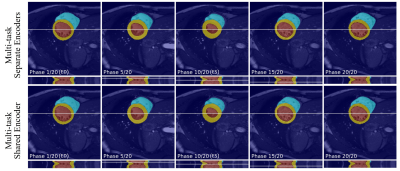
Figure 4. Visualization of segmentation of myocardium, left ventricular and right ventricular with our approach for a representative subject at multiple frames.