0709
Self-supervised contrastive learning for MR image reconstruction of cardiac CINE on accelerated cohorts1Medical Image and Data Analysis (MIDAS.lab), Department of Diagnostic and Interventional Radiology, University Hospital of Tuebingen, Tuebingen, Germany, 2Lab for AI in Medcine, Technical University of Munich, Munich, Germany, 3Department of Computing, Imperial College London, London, United Kingdom, 4Max Planck Institute for Intelligent Systems, Tuebingen, Germany
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence, Self-Supervised Learning, Image Reconstruction, Heart
Existing methods for deep-learning reconstruction require abundant fully-sampled images as labels, which is challenging or impractical in practice. To address this issue, we propose a self-supervised learning (SSL) approach that can be trained on subsampled data, avoiding the need for fully-sampled datasets. Specifically, we pre-train a feature extractor by contrastive learning as the first step. In the second step, the pre-trained feature extractor assists the self-supervised network during reconstruction by feature embedding. Results indicate that the proposed SSL method can effectively reconstruct cardiac CINE images without fully-sampled data. It outperforms existing SSL networks and shows comparable results to supervised learning.Introduction
With the development of deep learning methods for MR image reconstruction, the reconstruction speed and image quality have greatly improved. However, most of these methods1-4 require abundant fully-sampled high-resolution images for training, which is challenging or sometimes impractical to obtain considering the need for extended scan times, breath-holds, or acquisitions under free-movement.To address this issue, self-supervised learning (SSL) has been proposed5-7. However, their reachable acceleration rate is still limited and so far focused on 2D static imaging, which does not necessarily effectively capture spatial-temporal characteristics of motion-resolved data. Contrastive learning8 is an SSL method used to obtain general representations of the data. It has been studied as a regularizing loss on the reconstructed images7,9, but their feature embedding was not incorporated into the reconstruction network, limiting their potential.
In this work, we propose a self-supervised learning approach which can be trained on subsampled imaging data, i.e. avoiding the need for fully-sampled datasets. The reconstruction network is assisted by a feature extractor pre-trained with a contrastive learning strategy. To the best of our knowledge, this is the first study that combines contrastive learning and self-supervision for MR image reconstruction of undersampled motion-resolved cardiac CINE.
Methods
To leverage the representation learning ability of contrastive learning, our method is divided into two phases: contrastive feature learning and self-supervised reconstruction.Contrastive learning
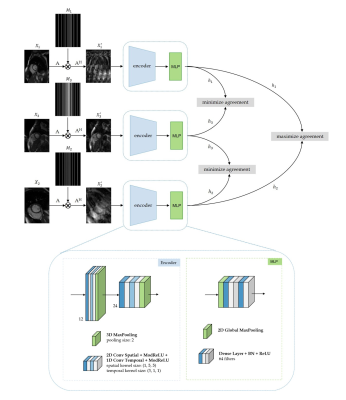
For contrastive learning, defining positive and negative pairs is essential for the network to know which data are similar or different, thus extracting reliable and valuable underlying image features. In the proposed contrastive learning (Fig.1), positive pairs are two similar undersampled images of the same subject. To create them, the fully-sampled image $$$\mathbf{x}_{1}$$$ is augmented (rotation, cropping) to obtain $$$\mathbf{x}_{2}$$$. The respective k-spaces $$$\mathbf{y}_{1}=\mathbf{Ax}_{1},\,\mathbf{y}_{2}=\mathbf{Ax}_{2}$$$ with encoding operator $$$\mathbf{A=MFS}$$$ consisting of Fourier transformation $$$\mathbf{F}$$$, coil sensitivity maps $$$\mathbf{S}$$$ and sampling mask $$$\mathbf{M}$$$, are then undersampled using two similar VISTA10 masks $$$(\mathbf{M}_{1},\,\mathbf{M}_{2})$$$ with the same acceleration rate but different seeds.
Positive pairs $$$\{\mathbf{x}_{1}^{'},\mathbf{x}_{2}^{'}\}$$$ are formed by the adjoint operations on $$$\mathbf{y}_{1}$$$ and $$$\mathbf{y}_{2}$$$, i.e. transforming the undersampled k-space back to image domain. While for this retrospective study, fully-sampled datasets were required, in prospective studies, this operation could be implemented by sampling a subject twice with two similar sampling masks, as we only require similarity in the image domain.
Negative pairs are two undersampled images $$$\{\mathbf{x}_{1}^{'},\mathbf{x}_{3}^{'}\}$$$ from two different subjects. To further enhance the dissimilarity, the difference in acceleration rate between two masks $$$(\mathbf{M}_{1},\,\mathbf{M}_{3})$$$ is $$$>5$$$. Similarly, $$$\{\mathbf{x}_{2}^{'},\mathbf{x}_{3}^{'}\}$$$ are also negative pairs.
The feature embedding network consists of an encoder (two stages of 2D+t complex-valued convolutions) and a multilayer perceptron (MLP). The MLP projects the extracted spatial-temporal features of $$$\mathbf{x}_{1}^{'}$$$, $$$\mathbf{x}_{2}^{'}$$$, $$$\mathbf{x}_{3}^{'}$$$ to a latent representation space $$$\mathbf{h}_{1}$$$, $$$\mathbf{h}_{2}$$$ and $$$\mathbf{h}_{3}$$$. During training, the contrastive loss maximizes the agreement between representations of positive pairs $$$(\{\mathbf{h}_{1},\mathbf{h}_{2}\})$$$ while repelling negative samples $$$(\{\mathbf{h}_{1},\mathbf{h}_{3}\},\,\{\mathbf{h}_{2},\mathbf{h}_{3}\})$$$.
Self-supervised reconstruction
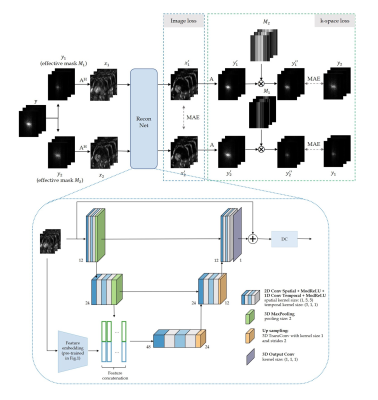
The proposed self-supervised reconstruction framework is shown in Fig.2. Undersampled k-space data $$$\mathbf{y}$$$ is further undersampled by VISTA10 to create two undersampled k-spaces $$$\mathbf{y}_{1}$$$ and $$$\mathbf{y}_{2}$$$, with their correspond sampling masks being $$$\mathbf{M}_{1}$$$ and $$$\mathbf{M}_{2}$$$. The respective undersampled images are fed into the reconstruction network, which is a physics-based network with only one iteration consisting of a complex-valued 2D+t UNet and data consistency (DC). The embedded features from the contrastive learning are concatenated in the UNet's bottleneck. The loss consists of image loss, MAE between the two output images $$$\mathbf{x}_{1}^{'}$$$ and $$$\mathbf{x}_{2}^{'}$$$, and k-space loss, for which the images are transformed into k-spaces $$$\mathbf{y}_{1}^{'},\mathbf{y}_{2}^{'}$$$ by $$$\mathbf{A}$$$ and then multiplied with the other's sampling mask $$$\mathbf{M}_{2},\mathbf{M}_{1}$$$. The MAE difference between reconstructed and sampled k-space points ($$$\mathbf{y}_{1}^{''}$$$ and $$$\mathbf{y}_{2}$$$, $$$\mathbf{y}_{2}^{''}$$$ and $$$\mathbf{y}_{1}$$$) is minimized.
2D cardiac CINE (bSSFP, TE/TR=1.06/2.12ms, $$$α=52°$$$, resolution=1.9x1.9mm2, slice thickness=8mm) was acquired on a 1.5T MRI in 129 subjects (38 healthy subjects and 91 patients). Data were split into 115 training and 14 test subjects. VISTA10 undersampling with random acceleration factor 2x to 16x is used in re-undersampling, resulting in an effective acceleration of 2x to 28x. The proposed method is compared to SSDU5 and a fully-supervised reconstruction. All comparisons use the same network architecture to study the pure impacts of training strategies.
Results and Discussion
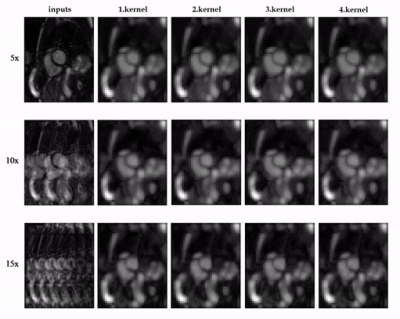
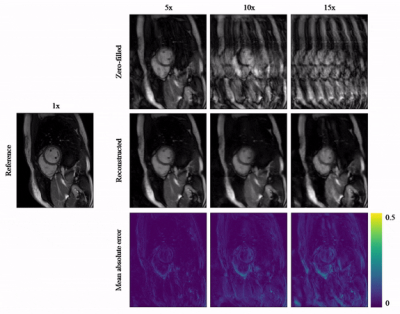
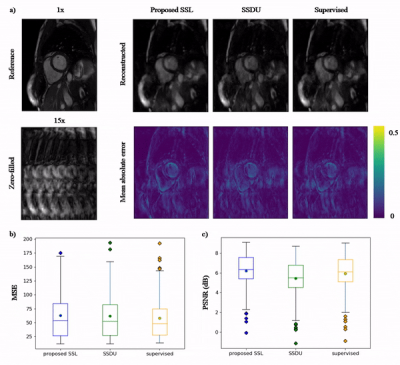
Animated Fig.3 shows the time-resolved learned features by contrastive learning for accelerations 5x, 10x and 15x. We observe a consistent and reliable feature extraction ability for the proposed contrastive learning strategy.Animated Fig.4 shows the time-resolved reconstructions by our self-supervised learning network for 5x, 10x and 15x retrospectively undersampled 2D CINE in one patient with active myocarditis. The comparison between SSDU5 and supervised learning is shown in Fig.5 (15x). Our proposed method effectively reconstructs images under different acceleration rates with good image quality. We observe suppressed aliasing artifacts in the myocardial area compared to SSDU5. Our method also outperforms SSDU5 in terms of PSNR, even showing a higher value and a more stable performance compared to supervised learning.
Conclusion
The pre-trained feature extractor can successfully extract features of clean images by contrastive training on undersampled images. With the extracted features embedded in the reconstruction network, the proposed self-supervised learning approach can efficiently reconstruct highly undersampled 2D cardiac CINE without fully-sampled data during training.Acknowledgements
S.G. and T.K. contributed equally.References
1. K. Hammernik et al., "Learning a variational network for reconstruction of accelerated MRI data", Magn Reson Med 2018; 79(6):2055-71.
2. H.Aggarwal et al., "MoDL: Model-Based Deep Learning Architecture for Inverse Problems", IEEE Trans Med Imaging 2019; 38(2):394-405.
3. J. Schlemper et al., " A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction", IEEE Trans Med Imaging 2018; 37(2):491-503.
4. T. Küstner et al., "CINENet: deep learning-based 3D cardiac CINE MRI reconstruction with multi-coil complex-valued 4D spatio-temporal convolutions", Sci Rep 2020; 10(1):13710.
5. B. Yaman et al., "Self-supervised learning of physics-guided reconstruction neural networks without fully sampled reference data", Magn Reson Med 2020; 84(6):3172-91.
6. B. Yaman et al., "Multi-mask self-supervised learning for physics-guided neural networks in highly accelerated magnetic resonance imaging", NMR Biomed 2022; e4798.
7. S. Wan et al., "PARCEL: Physics-based Unsupervised Contrastive Representation Learning for Multi-coil MR Imaging", IEEE/ACM Trans Comput Biol Bioinform 2022.
8. T. Chen et al., "A simple framework for contrastive learning of visual representations", International Conference on Machine Learning, PMLR 2020; 1597-1607.
9. Q. Yi et al., "Contrastive Learning for Local and Global Learning MRI Reconstruction", 2021.
10. R. Ahmad et al., "Variable density incoherent spatiotemporal acquisition (VISTA) for highly accelerated cardiac MRI", Magn Reson Med 2015; 74(5):1266-78.
Figures