0697
Fully Automated Segmentation of Brain and Scalp Blood Vessels on Multi-Parametric Magnetic Resonance Imaging Using Multi-view Cascaded Network1Department of Radiology, Suining Central Hospital, Suining, China, 2Radiology Department, Shenzhen University General Hospital and Shenzhen University Clinical Medical Academy, shenzhen, China, 3Medical AI Lab, School of Biomedical Engineering, Health Science Center, Shenzhen University, Shenzhen, China, 4Medical AI Lab, School of Biomedical Engineering, Health Science Center, Shenzhen University, shenzhen, China
Synopsis
Keywords: Vessels, Blood, brain blood vessel segmentation,Multi-Parametric,Multi-view
Accurate segmentation of blood vessels allow neurosurgical navigation and can help neurosurgeons accurate surgical and treatment plans. However, traditional blood vessel segmentation methods based on thresholds have limited performance. To solve problem, we proposed a cascaded DL network (MVPC-Net) that combines three refinements: multi-view learning, multi-parameter input, and a multi-view ensemble module-based strategy. The results of ablation experiments showed that, by adding all the refinements proposed, the performance of the baseline model improved from Dice similarity coefficient 0.865 to 0.922. Thus, our method can provide better segmentation of the brain, and scalp blood vessels and has potential for clinical application.Introduction
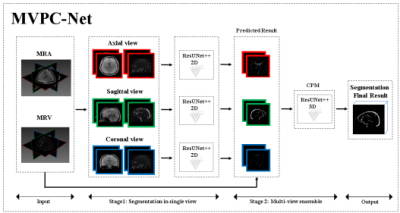
Blood vessel segmentation is important in neurosurgical navigation for clinical assessment of vascular diseases. The accurate segmentation of blood vessels is a useful reference source to allow doctors to improve surgical quality effectively1,2 . Compared with time-of-flight (TOF) magnetic resonance imaging (MRI), phase-contrast (PC) MRI has the advantages of better suppression of background (stationary) tissues and the ability to quantify and determine flow direction 3-7. Generally, PC MRI includes venography and angiography (MRV and MRA, respectively) parameter maps. However, manual segmentation of the brain and scalp vessels by radiologists is cumbersome. Thus, there is a need for automated vessel segmentation methods. Automatic segmentation of blood vessels has been studied for many years, and many algorithms based on traditional image processing methods have achieved success8-9. However, it is difficult to perform segmentation of blood vessels with different thicknesses using traditional image-processing methods, and these methods remain inadequate for segmentation of small blood vessels. Many automated models of deep convolutional neural networks have gradually been introduced and seem to show great potential 11-16. The above-mentioned segmentation method based on DL has the following limitations: First, it did not use multi-parameter MRI, and second, extracranial tissue was removed during pre-processing, although the vessel segmentation of these regions is also clinically required. In this study, to overcome the above-mentioned limitations of existing methods, we developed a multi-view multi-parameter cascaded network (MVPC-Net)( Figure 1) for blood vessel segmentation, based on PC MRI (MRV and MRA). In MVPC-Net, three two-dimensional (2D) ResUnet++with a ResNet encoder and Unet++ decoder were used for segmentation in three views, and a 3D ensemble module was introduced to merge the segmentation results of the three 2D networks.Material and Methods
This study protocol was approved by the Review Board of Shenzhen University General Hospital and 40 participants with drug-resistant epilepsy were selected. The MRI scans were performed on a Siemens 3.0 Tesla MR scanner with a 64-channel phased array head and neck coil. High-resolution 3D PC MRA was performed with the following parameters: repetition time = 48.9 ms; echo time = 8.2 ms; flip angle = 15°; acquisition matrix = 256 × 256, and slice thickness = 1 mm. High-resolution 3D PC MRV was performed using the same parameters, except for Velocity-encoding:10 cm/s. In the ablation experiments, improvements were gradually added to the base model based on 2D ResUnet++ in order to verify the validity of our proposed improvements, and the performance was recorded simultaneously. In the comparison experiment, We compared our approach with other existing deep-learning-based SOTA cerebrovascular segmentation studies.Results
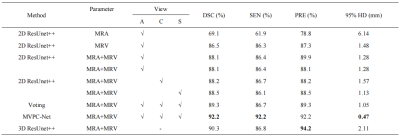
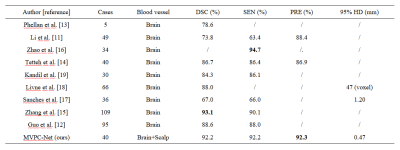
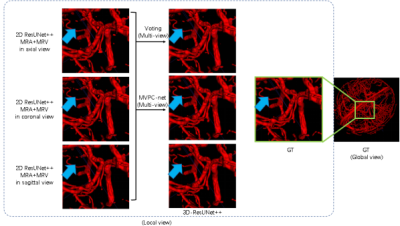
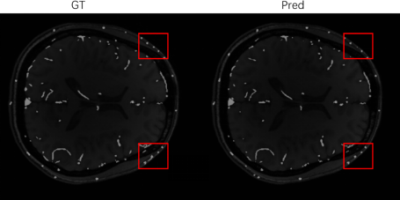
As the ablation experiment results shown in Table 1, by adding all the refinements proposed, the segmentation performance of the baseline model improved from Dice similarity coefficient 0.865 to 0.922. Visualizing the segmentation results, as shown in Figure 2, shows that using three perspectives for voting and using MVPC-Net with CPM can improve the details of blood vessel segmentation, with further marked improvement brought about by using CPM. As shown in Table 2, the segmentation results were substantially better than those of most of current studies in the literature. Compared to previous studies 15, our method has a slightly lower performance, but was not proven to be able to segment the scalp blood vessels, suggesting that this study was clinically more meaningful.Discussion
Our MVPC-Net achieved better vessel segmentation performance, competing with the results of previous research, and we improved the DSC from 86.5% to 92.2% as compared to the MRV-based ResUnet++ baseline. From the experimental results using multi-parameter input, it is clear that the imaging properties of MR images with different parameters are different in the same tissue. Because the direction of blood vessels in the brain is complex and anisotropic, pure 2D segmentation can only handle blood vessels coursing parallel to the plane. This may be why multi-view performance was better than single-view performance: multiple views complement each other so that the blood vessels coursing in all directions can be well segmented. Because there are some differences in the components of the brain, other studies 11-19 have generally not considered parts outside the brain, but have excluded them as interference components. However, our model performed well in the segmentation of blood vessels in the outer part of the brain, as shown in Figure 3. Therefore, we included this information in the input image. We suspect that, while regions outside of the brain may interfere with other components, such as the skull, the way it contains blood vessels may provide new learning opportunities for the model to achieve better performance.Conclusion
In conclusion, we proposed a novel cascaded network framework, MVPC-Net, and showed that it achieves high performance in head blood vessel segmentation. To a certain extent, our proposed method can initially solve the problem of segmenting small blood and low-contrast vessels. Simultaneously, it allowed better segmentation of the brain blood vessels and scalp blood vessels from the original image. It is hoped that the proposed method can provide effective 3D brain blood vessel segmentation for patients and assist neurosurgeons in preoperative pathway planning or formulating treatment plans for vessel diseases in future.Acknowledgements
No acknowledgement found.References
1. Moche, M., et al., Navigation concepts for MR image‐guided interventions. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine, 2008. 27(2): p. 276-291.
2. Sang, S., et al., Clinical long-term follow-up evaluation of functional neuronavigation in adult cerebral gliomas. World neurosurgery, 2018. 119: p. e262-e271.
3. Dumoulin, C.L., Phase contrast MR angiography techniques. Magnetic resonance imaging clinics of North America, 1995. 3(3): p. 399-411.
4. Huston 3rd, J. and R.L. Ehman, Comparison of time-of-flight and phase-contrast MR neuroangiographic techniques. Radiographics, 1993. 13(1): p. 5-19.
5. Pelc, N.J., et al., Fundamentals of flow and hemodynamics. Magnetic resonance imaging of the brain and spine, 2002. 3: p. 118-122.
6. Turski, P. and F. Korosec, Technical features and emerging clinical applications of phase-contrast magnetic resonance angiography. Neuroimaging Clin N Am, 1992. 2(4): p. 785-800.
7. Van Goethem, J., et al., Phase-contrast magnetic resonance angiography. JBR-BTR: Organe de la Societe Royale Belge de Radiologie (SRBR)= Orgaan van de Koninklijke Belgische Vereniging Voor Radiologie (KBVR), 2003. 86(6): p. 340-344.
8. Sahoo, P.K., S. Soltani, and A.K. Wong, A survey of thresholding techniques. Computer vision, graphics, and image processing, 1988. 41(2): p. 233-260.
9. Manniesing, R., M.A. Viergever, and W.J. Niessen, Vessel enhancing diffusion: A scale space representation of vessel structures. Medical image analysis, 2006. 10(6): p. 815-825.
10. Wilson, D.L. and J.A. Noble, An adaptive segmentation algorithm for time-of-flight MRA data. IEEE transactions on medical imaging, 1999. 18(10): p. 938-945.
11. Chen, L., et al. 3D intracranial artery segmentation using a convolutional autoencoder. in 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). 2017. IEEE.
12. Guo, X., et al., Cerebrovascular segmentation from TOF-MRA based on multiple-U-net with focal loss function. Computer Methods and Programs in Biomedicine, 2021. 202: p. 105998.
13. Phellan, R., et al., Vascular segmentation in TOF MRA images of the brain using a deep convolutional neural network, in Intravascular Imaging and Computer Assisted Stenting, and Large-Scale Annotation of Biomedical Data and Expert Label Synthesis. 2017, Springer. p. 39-46.
14.Tetteh, G., et al., Deepvesselnet: Vessel segmentation, centerline prediction, and bifurcation detection in 3-d angiographic volumes. Frontiers in Neuroscience, 2020: p. 1285.
15. Zhang, B., et al., Cerebrovascular segmentation from TOF-MRA using model-and data-driven method via sparse labels. Neurocomputing, 2020. 380: p. 162-179.
16. Zhao, F., et al., Semi-supervised cerebrovascular segmentation by hierarchical convolutional neural network. IEEE Access, 2018. 6: p. 67841-67852.
17. Sanches, P., et al. Cerebrovascular network segmentation of MRA images with deep learning. in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). 2019. IEEE.
18. Livne, M., et al., A U-Net deep learning framework for high performance vessel segmentation in patients with cerebrovascular disease. Frontiers in neuroscience, 2019. 13: p. 97.
19. Kandil, H., et al. Using 3-D CNNs and local blood flow information to segment cerebral vasculature. in 2018 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT). 2018. IEEE.
Figures