0688
3D diffusion MRI with twin-navigator-based GRASE for cortical gray matter time-dependency measurements in the human brain1Key Laboratory for Biomedical Engineering of Ministry of Education, Department of Biomedical Engineering, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, China, 2Siemens Healthineers Ltd, Hangzhou, China
Synopsis
Keywords: Diffusion/other diffusion imaging techniques, Brain
3D oscillating gradient sequences enable diffusion measurement at short diffusion-time (td), but it suffered from low resolution and low SNR in clinical systems, and thus, the td-dependency in complex structures, such as cortical gray matter, was not characterized. Here we proposed a twin-navigator-based 3D oscillating gradient diffusion-weighted gradient spin-echo sequence for high-resolution whole-brain td–dMRI acquisition. We demonstrated that different cortical regions exhibited distinct td-dependency patterns with different diffusion dispersion exponent (θ) based on the power-law. We found θ was greater than 0.5 with the highest dispersion in the pre- and post-central cortex and lowest value in the frontal region.Introduction
Oscillating gradient (OG)1 has been used to assess diffusion at short diffusion-time (td), and to infer about the tissue microstructures2-5. However, this technique suffered from low signal-to-noise (SNR) and low b-value due to the limited gradient strength on clinical scanners. We recently proposed a 3D oscillating gradient-prepared gradient spin-echo (OGprep-GRASE) sequence to target these problems6, 7. However, whole-brain high-resolution 3D dMRI remains challenging due to the phase errors from eddy current and motion, which is complicated by diffusion encoding in multishot acquisition.Here we proposed a multi-shot OGprep-GRASE equipped with a twin-navigator for 3D multi-shot whole-brain dMRI. Moreover, we accelerated the sequence with Controlled Aliasing in Parallel Imaging Results in Higher Acceleration (CAIPIRINHA)8 method to accelerate the acquisition. We compared the proposed 3D GRASE sequence with 2D EPI and quantified the td-dependency in cortical gray matter (GM) of the human brain on a 3T system, which was not characterized previously due to the limited resolution for these fine structures.
Methods
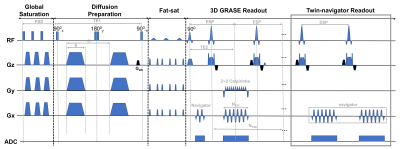
Pulse sequence: The twin-navigator-based 3D multi-shot OGprep-GRASE sequence consisted of five modules: global saturation, diffusion preparation, fat saturation, 3D GRASE readout, and twin-navigator readout (Figure 1).Data acquisition: Seven healthy volunteers were enrolled with IRB approval. All scans were performed on a 3T Siemens Prisma scanner with a 64-channel head coil. The 3D sequence parameters were: TR/TE1 (TE of the diffusion module)/TE2 (TE of the GRASE module) = 3000/86.68/49.8 ms, NEPI = 113, NTSE = 4, 12shots along the spin-echo encoding direction, and CAIPIRINHA accelerated factor = 4. Pulsed gradient (PG)-encodings at △eff = 20ms,30ms,40ms, and OG-encodings at frequencies of 25 Hz (2 cycles) and 50 Hz (4cycles) (effective td = 10ms and 5ms). The 2D-EPI were acquired with TR/TE = 18000/136 ms, NEPI = 132, and GRAPPA accelerated factor = 2, only with OG-encoding at frequencies of 25 Hz for comparison of SNR. We used one average for 3D-GRASE and 2 averages for 2D-EPI to ensure a similar scan time of approximately 10mins each td. The other parameters were kept the same: FOV = 220 × 200 mm2, voxel size = 1.5 × 1.5 × 1.5 mm3. b = 500 s/mm2 with 6 directions and two averages and four b0. We also acquired a 3D T1-weighted image for the segmentation of GM.
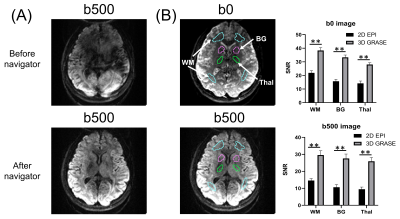
Data analysis: The acquired K-space data were reconstructed by CAIPIRINHA reconstruction9 and twin-navigator-based phase correction for 3D data. The SNR of both b0 and DWIs was calculated in representative gray and white matter regions that were manually delineated on b0 images of both 3D-GRASE and 2D-EPI data (Figure 2). The cortical and subcortical GM regions were segmented using the T1w image according to the AAL atlas10, which was co-registered to dMRI data. The ADC measurements from multi-td OG-dMRI and PG-dMRI data were obtained in each GM parcel to estimate the diffusion dispersion exponent (θ) based on the power-law model11: Dt =c×tθ +Dt0.
Results
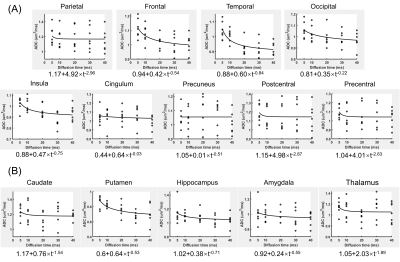
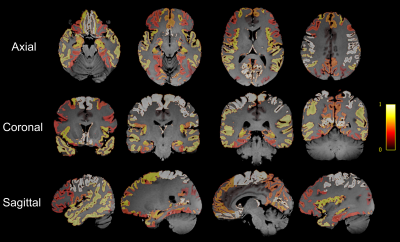
Quantitative comparison of SNR between the 2D-EPI and 3D-GRASE sequences (Figure 2) demonstrated that the SNR of the GRASE sequence was significantly (p<0.01) higher than the EPI sequence by approximately 1.75, 2.12, and 1.97 folds in the subcortical white matter, basal ganglia, and thalamus of both b0 and DWI.The td-dependent changes of ADC were displayed in several cortical and subcortical GM regions from the 3D GRASE sequence measured at different tds (Figure 3). Power-law analysis showed θ was greater than 0.5 in most ROIs except for the occipital and cingulum. Figure 4 mapped θ of all the GM regions on the T1 image, which showed the sensory cortex, such as the pre- and post-central regions had the highest dispersion and the high-order cortex such as frontal and temporal regions exhibited the lowest value.
Discussion and Conclusion
In this work, we proposed a twin-navigator-based 3D multishot OGprep-GRASE sequence for whole-brain dMRI acquisition on a clinical 3T system and investigated the td-dependent diffusivity in the cortical and subcortical GM regions. The twin-navigator approach effectively corrected the inter-shot phase errors and considerably improved SNR compared to 2D-EPI, which improved the td-dependency measurements. Besides, the current sequence is equipped with CAIPIRINHA to reduce the TE.Based on 3D GRASE measurements, we identified distinct td-dependency profiles in the human brain. The diffusion dispersion model showed the estimated θ > 0.5 in most GM regions, which was consistent with the recent report11, 12. Since higher θ indicated more organized microstructural organization, the current finding of higher θ in the sensory cortex and lower θ in the frontal and temporal cortex may indicate more complex organization in the high-order region compared to the sensory cortex.
Acknowledgements
This work is supported by the Ministry of Science and Technology of the People’s Republic of China (2018YFE0114600 and 2021ZD0200202), the National Natural Science Foundation of China (81971606 and 82122032), and the Science and Technology Department of Zhejiang Province (202006140 and 2022C03057).References
1. Schachter M, Does M, Anderson A, et al. Measurements of restricted diffusion using an oscillating gradient spin-echo sequence. Journal of magnetic resonance 2000;147(2):232-237.
2. Wu D, Martin LJ, Northington FJ, et al. Oscillating gradient diffusion MRI reveals unique microstructural information in normal and hypoxia‐ischemia injured mouse brains. Magnetic resonance in medicine 2014;72(5):1366-1374.
3. Ianuş A, Shemesh N, Alexander DC, et al. Double oscillating diffusion encoding and sensitivity to microscopic anisotropy. Magnetic resonance in medicine 2017;78(2):550-564.
4. Ianuş A, Jespersen SN, Duarte TS, et al. Accurate estimation of microscopic diffusion anisotropy and its time dependence in the mouse brain. Neuroimage 2018;183:934-949.
5. Arbabi A, Kai J, Khan AR, et al. Diffusion dispersion imaging: mapping oscillating gradient spin‐echo frequency dependence in the human brain. Magnetic resonance in medicine 2020;83(6):2197-2208.
6. Wu D, Liu D, Hsu YC, et al. Diffusion‐prepared 3D gradient spin‐echo sequence for improved oscillating gradient diffusion MRI. Magnetic resonance in medicine 2020;85(1):78-88.
7. Li H, Zu T, Hsu YC, et al. Inversion‐Recovery‐Prepared Oscillating Gradient Sequence Improves Diffusion‐Time Dependency Measurements in the Human Brain. Journal of Magnetic Resonance Imaging 2022.
8. Breuer F, Blaimer M, Griswold M, et al. Controlled aliasing in parallel imaging results in higher acceleration (CAIPIRINHA). Magnetom Flash 2012;1:2-7.
9. Qu P, Wang C, Shen GX. Discrepancy‐based adaptive regularization for GRAPPA reconstruction. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine 2006;24(1):248-255.
10. Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002;15(1):273-289.
11. Lee H-H, Papaioannou A, Novikov DS, et al. In vivo observation and biophysical interpretation of time-dependent diffusion in human cortical gray matter. Neuroimage 2020;222:117054.
12. Olesen JL, Østergaard L, Shemesh N, et al. Diffusion time dependence, power-law scaling, and exchange in gray matter. NeuroImage 2022;251:118976.
Figures