0675
Accelerated T2 Mapping with GS-Assisted Deep Translation of T1W Image Prior1School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3National Center for Supercomputing Applications, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 4Siemens Medical Solutions USA, Inc., Urbana, IL, United States, 5Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 6Radiology Department, The Fifth People's Hospital of Shanghai, Shanghai, China
Synopsis
Keywords: Quantitative Imaging, Machine Learning/Artificial Intelligence, T2 mapping
Deep learning (DL)-based methods have shown great potential for accelerating T2W imaging by using image prior generated from a companion T1W image. However, quantitative T2 mapping requires multiple T2W images acquired with multi-TE, creating practical problem for the use of DL for accelerated T2 mapping due to insufficient multi-TE training data. This work addresses this problem by using a generalized series model to map a T2W DL prior for one TE to multiple TEs, enabling effective use of T1W image prior for high-quality T2 mapping from sparsely sampled data. The method has been validated using experimental data, producing impressive results.Introduction
Quantitative T2 mapping requires the acquisition of multiple T2W data with different TEs, often leading to a long scan time. In practice, T2 mapping is usually accompanied by a fast T1W scan, which can be used as a constraint to reconstruct T2W images from limited k-space data to reduce scan time1-3. Recently, several deep learning (DL)-based methods have been proposed to use a deep network to translate the companion T1W image into a T2W prior for constrained reconstruction of the T2W image4-7. However, quantitative T2 mapping requires multiple T2W images acquired with multiple TEs, which creates a practical problem for the use of current DL methods for accelerated T2 mapping due to the lack of sufficient multi-TE training data. This work addresses this problem by using a generalized series (GS) model to map a T2W DL prior for one TE to multiple TEs, enabling effective use of T1W image prior for high-quality T2 mapping from sparsely sampled sensitivity-encoded data. The proposed method has been validated using experimental data, producing impressive results.Method
The proposed method, illustrated in Fig. 1, integrates DL-based image translation and GS-based reconstruction to generate T2W image priors for multiple TEs, which are then used for the estimation of coil sensitivity functions and reconstruction of T2 maps from sparsely sampled k-space data.Deep neural network to generate single-TE image prior
This work is focused on brain imaging. We used the Human Connectome Project (HCP) database9 to train a pix2pixGAN network10. After training, the network is capable of translating a T1W image into a T2W image prior for a single TE.
GS-based reconstruction to generate sensitivity-weighted multi-TE image priors
We represented the sensitivity-weighted T2W image $$$\rho_{\text{GS},c}(x,\text{TE})$$$ for the $$$c^{th}$$$ coil of a specific TE by:$$\hspace{26.2em}\rho_{\text{GS},c}(x,\text{TE})=\sum_{n=-N_c/2}^{N_c/2}b_{n,c}(\text{TE})\rho_{\text{ref}}(x)e^{i2\pi n\triangle kx},\hspace{26.2em}(1)$$where $$$\rho_{\text{ref}}(x)e^{i2\pi n\triangle kx}$$$ represents the new basis functions incorporating DL T2W prior (and tissue-based intensity matching). $$$b_{n,c}(\text{TE})$$$ are the GS coefficients that are adaptively determined for each individual coil and TE by fitting the GS model to $$$d_{c}(\text{TE})$$$, the measured data from the $$$c^{th}$$$ coil of TE. This is a linear problem that can be solved efficiently using the least squares fitting method.
After the $$$\widehat{b}_{n,c}(\text{TE})$$$ are determined, $$$\widehat{\rho}_{\text{GS},c}(x,\text{TE})$$$ provides an image prior (or initial reconstruction) for each TE. In other words, the GS model maps the DL translation prior to multiple TEs. In addition, from $$$\widehat{\rho}_{\text{GS},c}(x,\text{TE})$$$, for all the $$$c’\text{s}$$$, we can obtain the sensitivity function for each coil using the sum-of-squares (SoS) method, that is:$$\hspace{33.4em}\widehat{S}_{c}=\frac{\widehat{\rho}_{\text{GS},c}}{\widehat{\rho}_{\text{GS}}},\hspace{33.4em}(2)$$where $$$\widehat{\rho}_{\text{GS}}=\sqrt{\sum_{c=1}^C\widehat{\rho}_{\text{GS},c}(\widehat{\rho}_{\text{GS},c})^\text{H}}.$$$
Constrained image reconstruction incorporating the GS-based image priors
We incorporated the priors in the form of coil-combined multi-TE images $$$\widehat{\rho}_{\text{GS}}$$$, and sensitivity function $$$\widehat{S}$$$ by:$$\hspace{23em}\triangle\widehat{\rho}_n=\mathop{\arg\min}\limits_{\triangle\rho_n} ||d-\Omega F\widehat{S}(\widehat{\rho}_{\text{GS}} + \triangle\rho_{n})||_2^2 +\lambda ||W(\triangle\rho_{n})||_{1},\hspace{23em}(3)$$where $$$\triangle\rho_{n}$$$ represents the residual image features, $$$d$$$ the measured data, $$$\Omega$$$ the sampling operator, $$$F$$$ the Fourier operator and $$$W$$$ the sparsifying transform.
Results
The proposed method has been validated using experimental data acquired from a healthy volunteer and a tumor patient at 3T scanners (MAGNETOM Prisma and Skyra, Siemens Healthcare, Erlangen, Germany). Multi-TE T2W images were acquired using the TSE sequence with the following imaging parameters: TR = 11000ms, TEs = [12ms,70ms,128ms,186ms,256ms], FOV = 240x240x72mm, and matrix size = 256x256x24; T1W images were acquired using the MPRAGE sequence with the following parameters: TR/TE = 2400/2.1ms, FOV = 256x192x256mm, and matrix size = 256x192x256. The HCP dataset9 was used for model training, which included paired T1W and T2W (single TE) images acquired from 1200 subjects. Figure 2 demonstrates the capability of the proposed method in generating multi-TE T2W images. Figure 3 shows that the translated multi-TE T2W images enabled a better estimation of coil sensitivity as compared to the existing coil sensitivity estimation methods (e.g., ESPIRIT and sum-of-squares). The translated multi-TE T2W image priors and improved sensitivity functions yielded significantly better reconstruction performance of T2 map over three existing reconstruction methods, as shown in Fig. 4. We have also tested the proposed method on a patient data with brain tumor. As shown in Fig. 5, the proposed method produced a more accurate T2 map over the existing methods, with a better representation of the lesion.Conclusion
We presented a new method to integrate deep learning and GS modeling for accelerated T2 mapping. The proposed method translates a T1W image into T2W image priors for multiple TEs which are used for sensitivity estimation and reconstruction of T2W images from highly sparse data. The proposed method has been validated using experimental data, producing impressive results. The method may prove useful for MRI studies that involve quantitative T2 mapping.Acknowledgements
No acknowledgement found.References
1. Weizman L, Eldar Y C, Ben Bashat D. Reference‐based MRI. Medical physics. 2016;43(10): 5357-5369.
2. Ehrhardt M J, Betcke M M. Multicontrast MRI reconstruction with structure-guided total variation. SIAM Journal on Imaging Sciences. 2016;9(3): 1084-1106.
3. Song P, Weizman L, Mota J F C, et al. Coupled dictionary learning for multi-contrast MRI reconstruction. IEEE Transactions on Medical Imaging. 2019;39(3): 621-633.
4. Lyu Q, Shan H, Steber C, et al. Multi-contrast super-resolution MRI through a progressive network. IEEE Transactions on Medical Imaging. 2020;39(9): 2738-2749.
5. Dar S U H, Yurt M, Shahdloo M, et al. Prior-guided image reconstruction for accelerated multi-contrast MRI via generative adversarial networks. IEEE Journal of Selected Topics in Signal Processing. 2020;14(6): 1072-1087.
6. Xuan K, Xiang L, Huang X, et al. Multi-modal MRI reconstruction assisted with spatial alignment network. IEEE Transactions on Medical Imaging. 2022.
7. Meng Z, Guo R, Li Y, et al. Accelerating T2 mapping of the brain by integrating deep learning priors with low‐rank and sparse modeling. Magnetic Resonance in Medicine. 2021;85(3): 1455-1467.
8. Liang Z-P, Lauterbur P C. An efficient method for dynamic magnetic resonance imaging. IEEE Transactions on Medical Imaging. 1994;13(4): 677-686.
9. Van Essen D C, Smith S M, Barch D M, et al. The WU-Minn Human Connectome Project: An overview. Neuroimage. 2013;80:62-79.
10. Isola P, Zhu J Y, Zhou T, et al. Image-to-image translation with conditional adversarial networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017.
Figures
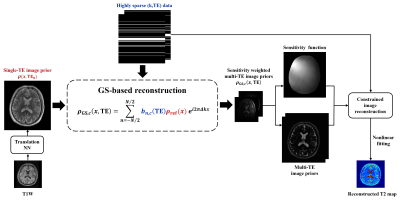
Figure 1: Overview of the proposed method that integrates DL-based image translation and generalized series (GS)-based reconstruction to generate T2W image priors for multiple TEs. The T2W priors are then used for the estimation of coil sensitivity functions and reconstruction of T2 maps from sparsely sampled k-space data.
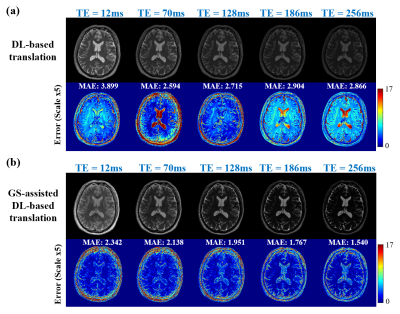
Figure 2: T2W image priors for multiple TEs generated using: (a) DL-based translation alone, and (b) GS-assisted DL-based translation. As can be seen, the proposed method significantly improved the T2W priors generated based on the companion T1W image and measured data.
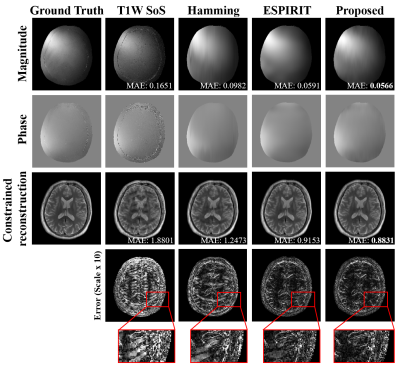
Figure 3: Illustration of the improved coil sensitivity estimation using the GS-based multi-TE image priors. Mean absolute error (MAE) of the sensitivity maps is shown in the magnitude image. As can be seen, the proposed method better estimates the sensitivity map, leading to an improved constrained image reconstruction in the following (R = 8 with 8 ACS lines).
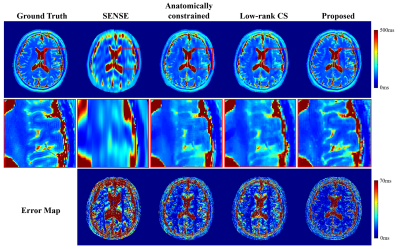
Figure 4: Comparison of the proposed method with three existing methods in recovering the T2 map of a healthy volunteer (R = 8). As shown, the proposed method produced a much better T2 map. The anatomically constrained reconstruction is implemented using an edge-preserving regularization, weighted L2. The weighting function is derived from the edge map of T1W image.
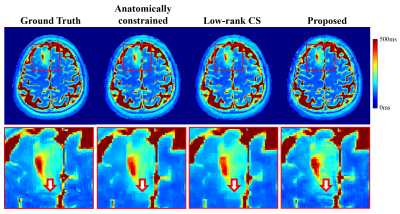
Figure 5: Comparison of the proposed method with two existing methods in recovering the T2 map of a tumor patient (R = 5). As shown, the proposed method produced a more accurate T2 map over the existing methods, with a better lesion boundary characterization as indicated by the arrow in the zoomed in images.