0638
De-noising of 4D real-time joint motion images using a convolutional neural network trained on static data
Laurel Hales1, Arjun Desai1, Valentina Mazzoli 1, Akshay Chaudhari1, and Feliks Kogan1
1Radiology, Stanford University, Stanford, CA, United States
1Radiology, Stanford University, Stanford, CA, United States
Synopsis
Keywords: Joints, Image Reconstruction, kinematic
We demonstrated the potential applying a noise removal machine learning network trained on images without motion, on a set of similar real-time images acquired with motion.Introduction
MSK disorders are a leading source of pain, but lack of knowledge about pain generators often leads to suboptimal patient outcomes. This is due in part to the fact that MSK pain maybe caused by maltracking or improper loading of joints. Most MRI imaging of joints is performed with the patient in a static position, without any load on the joint. The use of 4D real-time MR imaging could aid in studying disorders such as patellofemoral maltracking (PFM). Recent studies in this area have largely been limited to 2D single-slice acquisitions. But a single plane of motion is insufficient for the study motion in complex joints. The use of 3D acquisition would not only aid in diagnosis but would also make it possible to quantify the amount of maltracking present.Recent work by this group, demonstrated the potential of combining multiscale low-rank reconstruction with a 3D golden-ratio reordered cones acquisition to generate real-time 4D images in the knee.Figure 1 shows the early results in this area, and while the images are blurry, patellar motion is visible1.
The aim of this project is to continue this work and demonstrate that it is possible to remove some of those artifacts using a machine learning network trained on similar images acquired with the subject in a static position.
Materials and Methods
30 healthy subjects were scanned on a 3T whole-body scanner (GE SIGNA Premier) using a 16 channel receive-only phased-array flex coil, with subjects in a static position. Real-time 3D golden-ratio re-ordered cones data was acquired at multiple resolutions (FOV=16cm; matrix = 200x200x10,16, 128x128x10, 300x300x10, 400x400x10, 500x500x10, FA = 15; Slice Thickness = 3; TR/TE = 6.4/0.3 ms; total scan time ranged from 14 to 51 seconds.) Two subjects were scanned during slow contraction of their quadriceps muscle at a rate of 1 hertz. In these cases, a real-time 2D single-slice, single-shot fast-spin-echo (ssfse) sequence was also acquired for reference (FOV = 16.0 cm; matrix = 200x160x1;Slice Thickness = 1 cm; TR/TE = 305/52 ms; FA = 90; Temporal resolution ~305 ms). All cones images were reconstructed with a temporal resolution of ~500ms.Reconstruction was performed as previously described using a multiscale low-rank (MSLR) reconstruction algorithm with a lambda value of 1e-6 and block widths of 32, 64, and 1282. For comparison, a gridding and density compensation algorithm was used to create a fully sampled 3D image from the static data.
4D static images were used to train the machine learning network. These datasets were divided by participant into 42 training datasets, 6 validation datasets, and 6 test datasets. Each dataset consisted of 28 to 100 separate 3D volumes, one for each 500ms. Figure 2 shows a sample slice at sample timepoints of one of these datasets.
We built our network using the meddlr library3, and used the mateuszuda brain-segmentation architecture4. Each of the data was divided into patches of 128x128x1 pixels with a stride of 64x64x1. With a learning rate of 1e-4, batchsize of 32. The model was only trained for 4 epochs (about 10,000 iterations) since the focus of this project was to test the ability to apply a network to dynamic data and not create a fully trained noise removal network. This network was then applied to the data acquired with the subject in motion.
Results
Figure 3 shows the training and validation results of the network. The resulting images from the dynamic test set are shown in Figure 4. Figure 5 shows a gif of of a single slice of a knee during a quad flexDiscussion
Because there is no target data for the dynamic test-set, it is difficult to qualitatively measure the success of the method however, the noise level in the images is visibly reduced and This work demonstrated the feasibility that using a network trained on static data, could be applied to data with small amounts of motion, to remove noise and some artifacts.Current 2D approaches can image joint motion at frame rates of ~4 Hz but are limited to single slice acquisitions. 3D approaches have been limited to frame rates <0.5 Hz and imaging of slow joint motion that may not be sensitive to maltracking. This approach shows promise to bridge this gap. It was able to achieve 3D images with 12 slices and a temporal resolution of 500 ms. Further, the golden angle sampling utilized in the cones acquisition allows for retrospective rebinning to higher or lower temporal resolution depending on data quality and needs.
Further work is need to fine-tune the network. However, early indications are strong that we will be able to use a network that is trained on real-time images of a participant in a static position to remove noise and some under-sampling artifacts from real-time images of a participant in motion. Further, it maybe possible to acquire a static scan for each new subject in order to retrain the network to enable a subject specific improvement in network performance for motion data denoising. Recent work has also shown that fine tuning of the existing MRI protocol may also be effective in removing much of the noise and further improving the image quality.
Acknowledgements
Research funded by grants GE healthcare and nih R01AR079431 and R21EB030180
References
- Hales, L. and Kogan, F, “Three-Dimensional real-time dynamic knee MRI using 3D cones with a multiscale low-rank reconstruction” Proceedings of ISMRM, 2022
- Ong, F., Zhu, X., Cheng, J. Y., Johnson, K. M., Larson, P. E., Vasanawala, S. S., & Lustig, M. (2020). Extreme MRI: Large‐scale volumetric dynamic imaging from continuous non‐gated acquisitions. Magnetic resonance in medicine, 84(4), 1763-1780
- Meddlr Desai, A. D., Ozturkler, B. M., Sandino, C. M., Vasanawala, S., Hargreaves, B. A., Re, C. M., … Chaudhari, A. S. (2021). Noise2Recon: A Semi-Supervised Framework for Joint MRI Reconstruction and Denoising. arXiv preprint arXiv:2110. 00075.
- Buda, Mateusz, Ashirbani Saha, and Maciej A. Mazurowski. "Association of genomic subtypes of lower-grade gliomas with shape features automatically extracted by a deep learning algorithm." Computers in biology and medicine 109 (2019): 218-225.
Figures
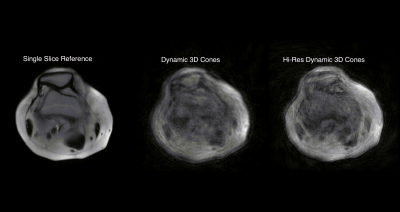
A single slice of a 4D data sent showing the potential of combining multi-scale low-rank reconstruction with a 3D golden-ratio reordered cones acquisition to generate real-time 4D images
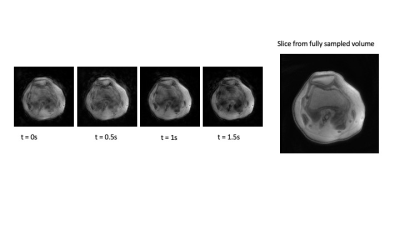
A representative slice shown at representative timepoints compared to the target fully sampled image.
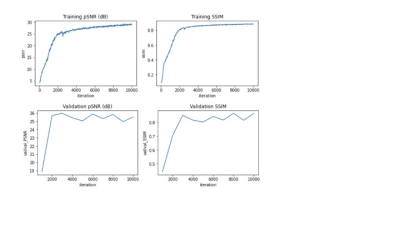
training and validation results. The final training loss was 0.01, with a total training MSE of 28.
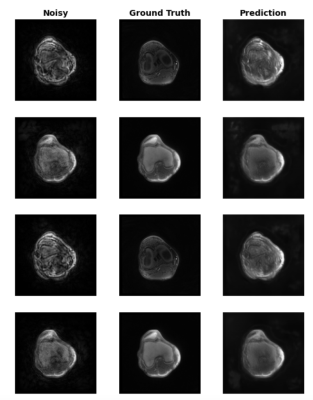
Individual slices and timepoints of dynamic data after noise removal
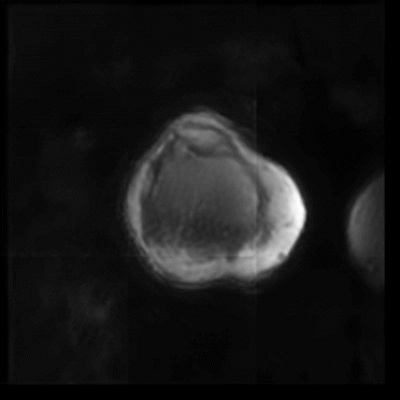
All time points of a representative slice of a real-time 4D knee with patellar motion.
DOI: https://doi.org/10.58530/2023/0638