0637
Self-supervised T1 Mapping from Two Variable Flip Angle Images without Requiring Ground Truth T1 Maps1Radiology, Stanford University, Stanford, CA, United States, 2Radiology, University of California San Diego, San Diego, CA, United States
Synopsis
Keywords: Cartilage, Cartilage, supervised learning
We propose a self-supervised learning method that derives T1 map from a reduced number of variable flip angle images without requiring ground truth maps, aimed at minimizing data acquisition efforts for obtaining training and testing data.Introduction
With the development of deep learning techniques, quantitative parametric T1 map can be derived from a reduced number of T1-weighted images [1]. However, to train a supervised learning model, full sets of 'weighted' images are still needed for the generation of ground truth parametric maps. In this study, we develop a self-supervised learning method to predict quantitative T1 maps from two variable flip angle (VFA) images without requiring ground truth maps, aimed at minimizing efforts for acquiring both training and testing data.Methods
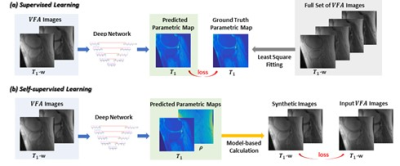
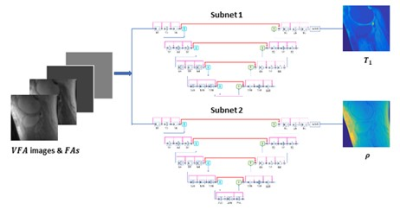
To mitigate the need for ground truth T1 maps in variable flip angle T1 mapping, self-supervised learning [2] was employed. At every iteration in training, the loss was calculated as the difference, not between the predicted T1 maps and the ground truth maps, but between the input images and the images synthesized from the predicted parametric maps. The comparison of self-supervised learning and supervised learning is illustrated in Figure 1.For self-supervised mapping, we designed a multi-output convolutional network, since PD (Proton Density) maps also need to be estimated (for image synthesis and loss calculation). The network predicted T1 and PD maps via parallel subnets (Figure 2), which were densely connected hierarchical convolutional networks, SAT-Nets [3-7].
A unique design of the proposed deep learning method was to include imaging parameters as additional network input. For every slice, not only were VFA images used as input, but also images that provided the values of flip angles at every pixel.
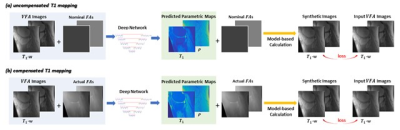
Separate models were established for T1 mapping with or without taking B1 inhomogeneities into compensation. In the uncompensated T1 mapping model (that ignored B1 inhomogeneities), the nominal/prescribed flip angles were incorporated as additional network input, facilitating the use of physical parameters as a priori information. In the revised compensated T1 mapping model (that considered B1 inhomogeneities), the actual flip angles (i.e., the combination of nominal flip angles and B1 map) were included as network input. Both deep learning models are illustrated in Figure 3.
With IRB approval and informed patient consent, variable flip angle images of the knee were acquired from 59 subjects. For every slice, four VFA images were acquired using an ultrashort echo time (UTE) cones sequence with flip angles of 5°, 10°, 20°, and 30° respectively, echo time of 32 µs, and time of repetition of 20 ms [8]. In addition, B1 map was measured using the actual flip angle method [9].
A total of 1224 two-dimensional images of the knee obtained from 59 subjects were used to train and test deep learning models with six-fold cross-validation applied. Out of the four VFA images, only two images acquired with flip angles of 10° and 20° were used as the network input.
A loss function was employed, which was defined as a combination of L1 norm and structural similarity index (SSIM): loss = L1+5*Lssim [3]. In every iteration, the L1-SSIM loss between synthesized images and input images was backpropagated, and network parameters were updated using the Adam algorithm with alpha of 0.001, beta1 of 0.89, beta2 of 0.89, and e of 10^-8.
In testing, every predicted T1 map was evaluated against the ground truth map (obtained from four VFA images). Quantitative evaluation was performed on the whole image as well as within the regions of interest (ROI), which was the cartilage region manually segmented.
Results
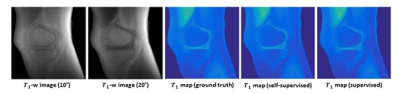
Uncompensated T1 mappingUsing uncompensated T1 mapping models, T1 maps were predicted from two VFA images and nominal flip angles (10° and 20°) with high fidelity. The self-supervised learning results were similar to supervised learning outcomes. A representative case is shown in Figure 4.
In uncompensated T1 mapping, we got a correlation coefficient of 0.9930 ± 0.0053, of 0.0311 ± 0.0027, and SSIM of 0.9548 ± 0.0044within the whole leg; a correlation coefficient of 0.9967 ± 0.0011, of 0.0386 ± 0.0103, and SSIM of 0.9999 ± 0.0000within the ROI in cartilage.
Compensated T1 mapping
Using compensated T1 mapping models, T1 maps were predicted from two VFA images and actual flip angles with high fidelity. An example is shown in Figure 5. Using actual flip angles (rather than B1 map) as network input helped to improve the prediction accuracy. This confirmed the benefit of incorporating imaging parameters as network input.
In compensated T1 mapping, we got a correlation coefficient of 0.9905 ± 0.0055, L1 of 0.0567 ± 0.0161, and SSIM of 0.9406 ± 0.0072 within the whole leg; a correlation coefficient of 0.9956 ± 0.0017, L1 of 0.0582 ± 0.0293, and SSIM of 0.9998 ± 0.0002 within the ROI in cartilage.
Conclusions
We propose a self-supervised learning method that derives T1 map from a reduced number of variable flip angle images without requiring ground truth maps, minimizing data acquisition efforts for obtaining training and testing data.Acknowledgements
The research was supported by National Institute of Health: NIH R01EB009690, NIH R01 EB026136, NIH R01DK117354 and GE Healthcare.References
1. Wu Y, et al. Accelerating quantitative MR imaging with the incorporation of B1 compensation using deep learning. Magnetic Resonance Imaging. 2020 Oct 1;72:78-86.
2. Senouf, Ortal, et al. "Self-supervised learning of inverse problem solvers in medical imaging." Domain adaptation and representation transfer and medical image learning with less labels and imperfect data. Springer, Cham, 2019. 111-119.
3. Y. Wu, et al, "Self-attention convolutional neural network for improved MR image reconstruction," Information Sciences, vol. 490, pp. 317-328, 2019.
4. Wu, Yan, et al. "Deciphering tissue relaxation parameters from a single MR image using deep learning." Medical Imaging 2020: Computer-Aided Diagnosis. Vol. 11314. International Society for Optics and Photonics, 2020.
5. Wu, Yan, et al. "Quantitative Parametric Mapping of Tissues Properties from Standard Magnetic Resonance Imaging Enabled by Deep Learning." arXiv preprint arXiv:2108.04912 (2021).
6. Wu Y, et al. Deriving new soft tissue contrasts from conventional MR images using deep learning. Magnetic Resonance Imaging. 2020 Dec 1;74:121-7.
7. Wu, Yan, et al. "Deep Learning-Based Water-Fat Separation from Dual-Echo Chemical Shift-Encoded Imaging." Bioengineering 9.10 (2022): 579.
8. YJ. Ma, et al, "3D adiabatic T1Ïprepared ultrashort echo time cones sequence for whole knee imaging," Magnetic Resonance in Medicine, vol. 80, pp. 1429-1439, 2018.
9. Yarnykh and V. Yarnykh, "Actual flip-angle imaging in the pulsed steady state: A method for rapid three-dimensional mapping of the transmitted radiofrequency field," Magnetic Resonance in Medicine, vol. 57, pp. 192-200, 2007.
Figures