0621
Layer-specific functional connectivity with 3D VAPER fMRI1Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Section on Functional Imaging Methods, NIMH, NIH, Bethesda, MD, United States, 3Children's National Hospital, Washington DC, DC, United States, 4Functional MRI Core, NIMH, NIH, Bethesda, MD, United States
Synopsis
Keywords: Brain Connectivity, fMRI, fMRI connectivity
We introduced a whole-brain 3D VAPER sequence tool and analysis approach for layer-specific resting state fMRI. It allows investigation of the directional connectivity between brain areas and determine whether any given connection is better described as predominantly feedforward vs. feedback driven. To exemplify this, we demonstrated that different directions of the same connection within the default mode network (mPFC <-> PCC/Parietal region) lead to different laminar correlation profile, suggesting different projection types (feedforward/feedback).Introduction
The layer-specific signals of functional connectivity have been poorly investigated, mainly due to coverage limitation enforced in high-resolution layer-fMRI imaging methods1. Recently, we developed a whole-brain layer-specific imaging sequence tool in humans2,3, for achieving fMRI at high resolution (0.8mm-isotropic), high specificity (not biased toward unspecific vein signals as BOLD), high sensitivity (robust measurement at layer-level resolution), high spatial accuracy (layer fMRI analysis in native fMRI space to avoid blurring and errors arising from registration), whole brain coverage, and enabling layer fMRI to acquire more flexible connectivity-based experiment designs. In order to illustrate its applicability of layer-specific functional connectivity research, we collect whole-brain submillimeter image data during resting state and investigate the involvement of the cortical layers in the maintenance of the resting-state dynamics, with here focusing on the default mode network.Methods
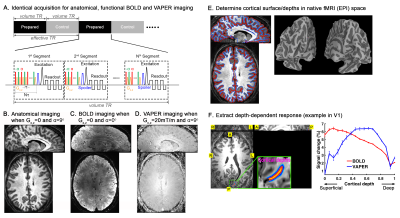
The experiments were performed on a Siemens MAGNETOM 7T scanner. For functional measurement, we acquired an integrated blood volume and perfusion (VAPER) weighted signals using VAPER-3D-EPI2 (Fig. 1). For anatomical reference, we incorporated magnetization transfer (MT) weighting3 with 3D-EPI using an identical sequence design as functional imaging, except for switching off the gradients in VAPER preparation (details in Fig. 1). Identical acquisition parameters were used for functional and anatomical imaging: volume TR = 5.898s, 96 slices (oversample additional 8.3%), excitation flip angle 20°, resolution 0.8-mm isotropic, partial-Fourier of 7/8 in both phase encoding directions, CAIPI 3x2 (kz shift 1, shot-selective). During functional runs, we instructed the participants to keep their heads still and not fall asleep. Due to the low sensitivity at submillimeter resolution, each resting state run lasted for 30 min and was repeated one more time in each session.According to the expectations of a canonical microcircuit4, feedforward driven input terminates mostly in middle cortical layers, while feedback input terminates in superficial and/or deep layers. With the correlation-value laminar profile for each network, this simplified layer-model allows us to develop binary classification algorithms (k-means and seed-based analysis) that determine whether any given connection is better described as predominantly feedforward driven vs. predominantly feedback driven.
Results
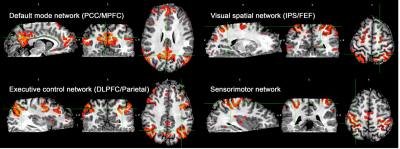
Fig. 1 shows the VAPER/MT-3D-EPI sequence design and the anatomical, functional BOLD and VAPER images it acquired for whole-brain at 0.8mm resolution. As the acquisition is identical, functional and anatomical images match each other during acquisition. This facilitates the determination of cortical depths in native fMRI space, obviating the need for distortion correction and registration, which will boost the accuracy of localizing high-resolution signal to different cortical layers.To demonstrate that the whole-brain 0.8-mm isotropic VAPER data has sufficient sensitivity for functional connectivity analysis, we used ICA to extract the common functional networks from the resting state VAPER data as in Fig. 2. To improve the TSNR, we used smoothing inside GM which was replaced with a layer-specific smoothing in later laminar analysis.
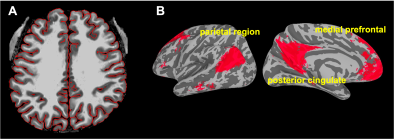
Next, we used the default mode network as an example to show how whole-brain high-resolution VAPER datasets can be used for layer-dependent connectivity analyses. At the individual level, we determined cortical depths on the anatomical EPI images (Fig. 3A) and then projected the functional resting-state data to the cortical surface at each cortical depth. To minimize the resolution loss during surface projection, we upsampled the functional volume data by a factor of 5 before projection and increased the vertex density (refinement iteration of 1)5. Then we applied the transformation from each individual to the group averaged cortical surface for the data at each cortical depth. This layer-specific surface registration will not cause any data blurring across cortical depths. Fig. 3B shows the group (N=12) averaged default mode network regions at the middle cortical depth.
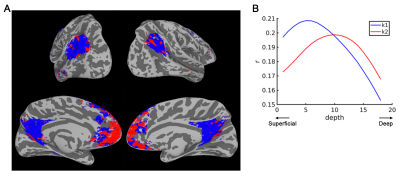
Then within the group-averaged default mode network, we applied k-means to cluster areas according to the similarity in their correlation-value laminar profile. The resulting cluster distribution (Fig. 4A) and the cluster-mean laminar profiles (Fig. 4B) shows that, the medial prefrontal cortex (mPFC) is mainly feedforward-driven (correlation peaks at middle cortical depths) while the inputs to parietal region and posterior cingulate (PCC) is predominantly feedback-driven (correlation peaks at superficial cortical depths).
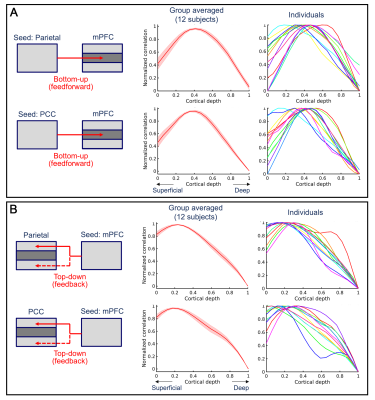
Last, we used a seed-based analysis to investigate the connectivity direction within default mode network. At each time, we used one of the three nodes (mPFC, parietal gion, PCC) as the seed ROI and extracted the time series of all voxels within the ROI. Next, for each voxel at different cortical depths in the other two nodes of DMN, the mean positive correlation to all seed voxels was computed. Fig. 5 shows that, when using PCC or parietal region as the seed, its correlation to mPFC peaks at middle cortical depths. While using mPFC as the seed, the correlation to PCC and parietal region peaks at superficial and/or deep cortical depths. It suggests that mPFC receives feedforward inputs from parietal region and PCC while parietal region and PCC receives feedback inputs from mPFC.
Conclusions
We introduced a whole-brain 3D-VAPER sequence tool for layer-specific resting-state fMRI. It allows investigation of the directional connectivity between brain areas. To exemplify this, we demonstrated that different directions of the same connection within the default mode network (mPFC <-> PCC/Parietal region) lead to different laminar correlation profile, suggesting different projection types (feedforward/feedback).Acknowledgements
P. Bandettini and B. Sutton are joint senior authors of this work. Part of this research was supported by the NIMH Intramural Research Program (ZIAMH002783). We thank Benedikt A. Poser for contributions to the 3D-EPI which was used in our sequence of VB17 version.References
1. Huber, L. et al. Layer-dependent functional connectivity methods. Prog Neurobiol 101835 (2020) doi:10.1016/j.pneurobio.2020.101835.
2. Chai, Y., Li, L., Huber, L., Poser, B. A. & Bandettini, P. A. Integrated VASO and perfusion contrast: A new tool for laminar functional MRI. Neuroimage 207, (2020).
3. Chai, Y. et al. Magnetization transfer weighted EPI facilitates cortical depth determination in native fMRI space. Neuroimage 242, (2021).
4. Felleman, D. J. & van Essen, D. C. Distributed hierarchical processing in the primate cerebral cortex. Cerebral Cortex 1, 1–47 (1991).
5. Wang, J., Nasr, S., Roe, A. W. & Polimeni, J. R. Critical factors in achieving fine‐scale functional MRI: Removing sources of inadvertent spatial smoothing. Hum Brain Mapp (2022) doi:10.1002/hbm.25867.
Figures